diff --git a/README.md b/README.md
index 61df072..1adb444 100644
--- a/README.md
+++ b/README.md
@@ -15,18 +15,26 @@ and allows to perform it in the differential comparison mode, or as a time-serie
As input, DIMet accepts three types of measures: a) isotopologues’ contributions, b) fractional contributions (also known as mean enrichment), c) full metabolites’ abundances.
DIMet also offers a _pathway-based omics integration_ through **Metabolograms**.
+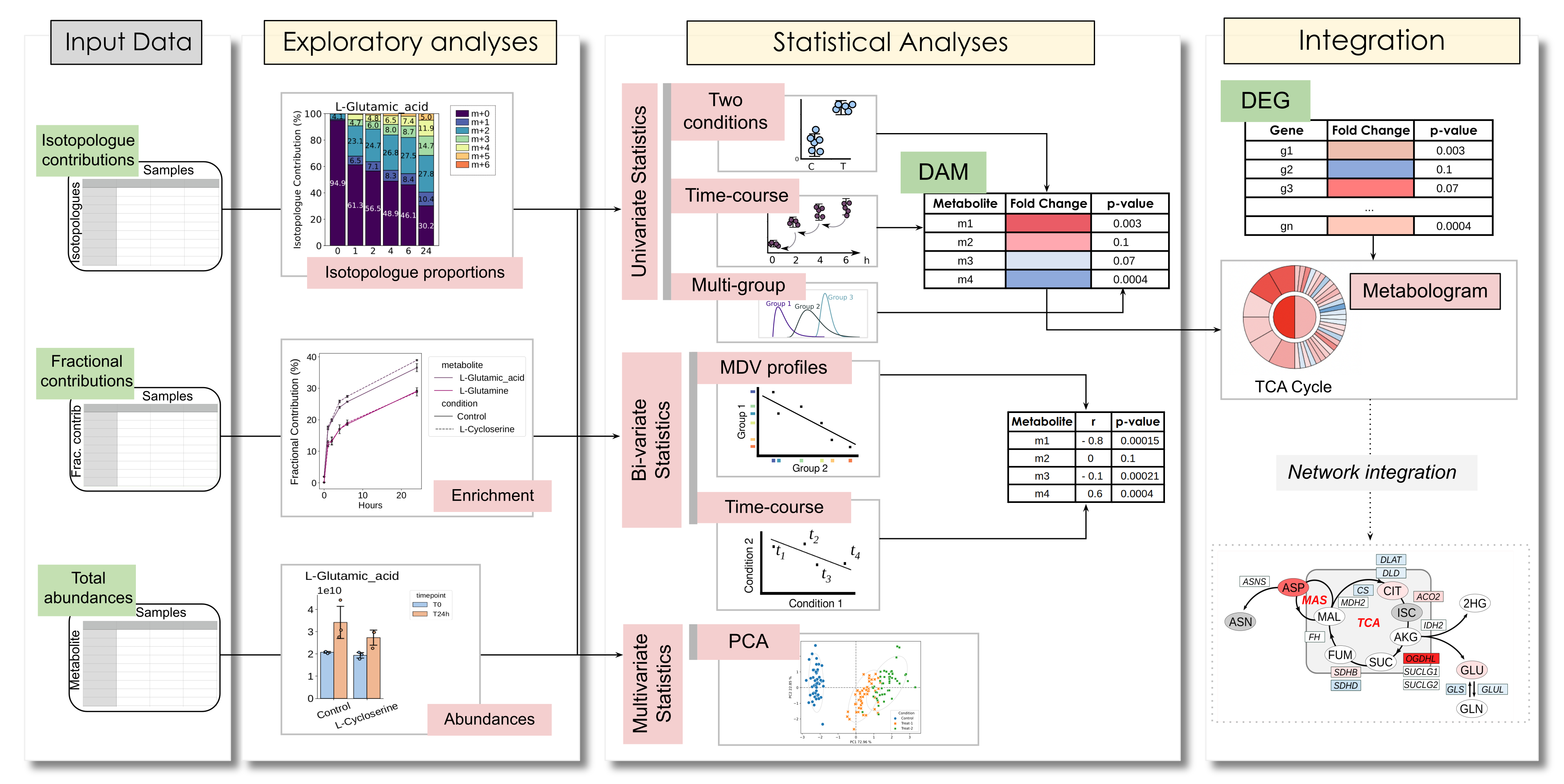 +DIMet functionalities (Galvis J., et al. Bioinformatics, 2024).
+
+
_Note_: DIMet is intended for downstream analysis of tracer metabolomics data that has been corrected for the presence of natural isotopologues.
_Formatting and normalisation helper_: scripts for formatting and normalization are provided in [TraceGroomer](https://github.com/cbib/TraceGroomer).
-_New_: DIMet is now available in **Galaxy**: Visit https://usegalaxy.eu/ to access the user-friendly interface of our tool.
+DIMet is available in **Galaxy Europe** (https://usegalaxy.eu/) and **Galaxy Workflow4Metabolomics** (https://workflow4metabolomics.usegalaxy.fr/), providing free access to the user-friendly interface of our tool.
> [!IMPORTANT]
> When using DIMet, please cite:
>
> Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An open-source tool for Differential analysis of targeted Isotope-labeled Metabolomics data. _Bioinformatics_ 2024; btae282. [https://doi.org/10.1093/bioinformatics/btae282](https://doi.org/10.1093/bioinformatics/btae282)
---------
+
+
+
+
+------
# Installing DIMet
+DIMet functionalities (Galvis J., et al. Bioinformatics, 2024).
+
+
_Note_: DIMet is intended for downstream analysis of tracer metabolomics data that has been corrected for the presence of natural isotopologues.
_Formatting and normalisation helper_: scripts for formatting and normalization are provided in [TraceGroomer](https://github.com/cbib/TraceGroomer).
-_New_: DIMet is now available in **Galaxy**: Visit https://usegalaxy.eu/ to access the user-friendly interface of our tool.
+DIMet is available in **Galaxy Europe** (https://usegalaxy.eu/) and **Galaxy Workflow4Metabolomics** (https://workflow4metabolomics.usegalaxy.fr/), providing free access to the user-friendly interface of our tool.
> [!IMPORTANT]
> When using DIMet, please cite:
>
> Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An open-source tool for Differential analysis of targeted Isotope-labeled Metabolomics data. _Bioinformatics_ 2024; btae282. [https://doi.org/10.1093/bioinformatics/btae282](https://doi.org/10.1093/bioinformatics/btae282)
---------
+
+
+
+
+------
# Installing DIMet
 +DIMet functionalities (Galvis J., et al. Bioinformatics, 2024).
+
+
_Note_: DIMet is intended for downstream analysis of tracer metabolomics data that has been corrected for the presence of natural isotopologues.
_Formatting and normalisation helper_: scripts for formatting and normalization are provided in [TraceGroomer](https://github.com/cbib/TraceGroomer).
-_New_: DIMet is now available in **Galaxy**: Visit https://usegalaxy.eu/ to access the user-friendly interface of our tool.
+DIMet is available in **Galaxy Europe** (https://usegalaxy.eu/) and **Galaxy Workflow4Metabolomics** (https://workflow4metabolomics.usegalaxy.fr/), providing free access to the user-friendly interface of our tool.
> [!IMPORTANT]
> When using DIMet, please cite:
>
> Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An open-source tool for Differential analysis of targeted Isotope-labeled Metabolomics data. _Bioinformatics_ 2024; btae282. [https://doi.org/10.1093/bioinformatics/btae282](https://doi.org/10.1093/bioinformatics/btae282)
---------
+
+
+
+
+------
# Installing DIMet
+DIMet functionalities (Galvis J., et al. Bioinformatics, 2024).
+
+
_Note_: DIMet is intended for downstream analysis of tracer metabolomics data that has been corrected for the presence of natural isotopologues.
_Formatting and normalisation helper_: scripts for formatting and normalization are provided in [TraceGroomer](https://github.com/cbib/TraceGroomer).
-_New_: DIMet is now available in **Galaxy**: Visit https://usegalaxy.eu/ to access the user-friendly interface of our tool.
+DIMet is available in **Galaxy Europe** (https://usegalaxy.eu/) and **Galaxy Workflow4Metabolomics** (https://workflow4metabolomics.usegalaxy.fr/), providing free access to the user-friendly interface of our tool.
> [!IMPORTANT]
> When using DIMet, please cite:
>
> Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An open-source tool for Differential analysis of targeted Isotope-labeled Metabolomics data. _Bioinformatics_ 2024; btae282. [https://doi.org/10.1093/bioinformatics/btae282](https://doi.org/10.1093/bioinformatics/btae282)
---------
+
+
+
+
+------
# Installing DIMet