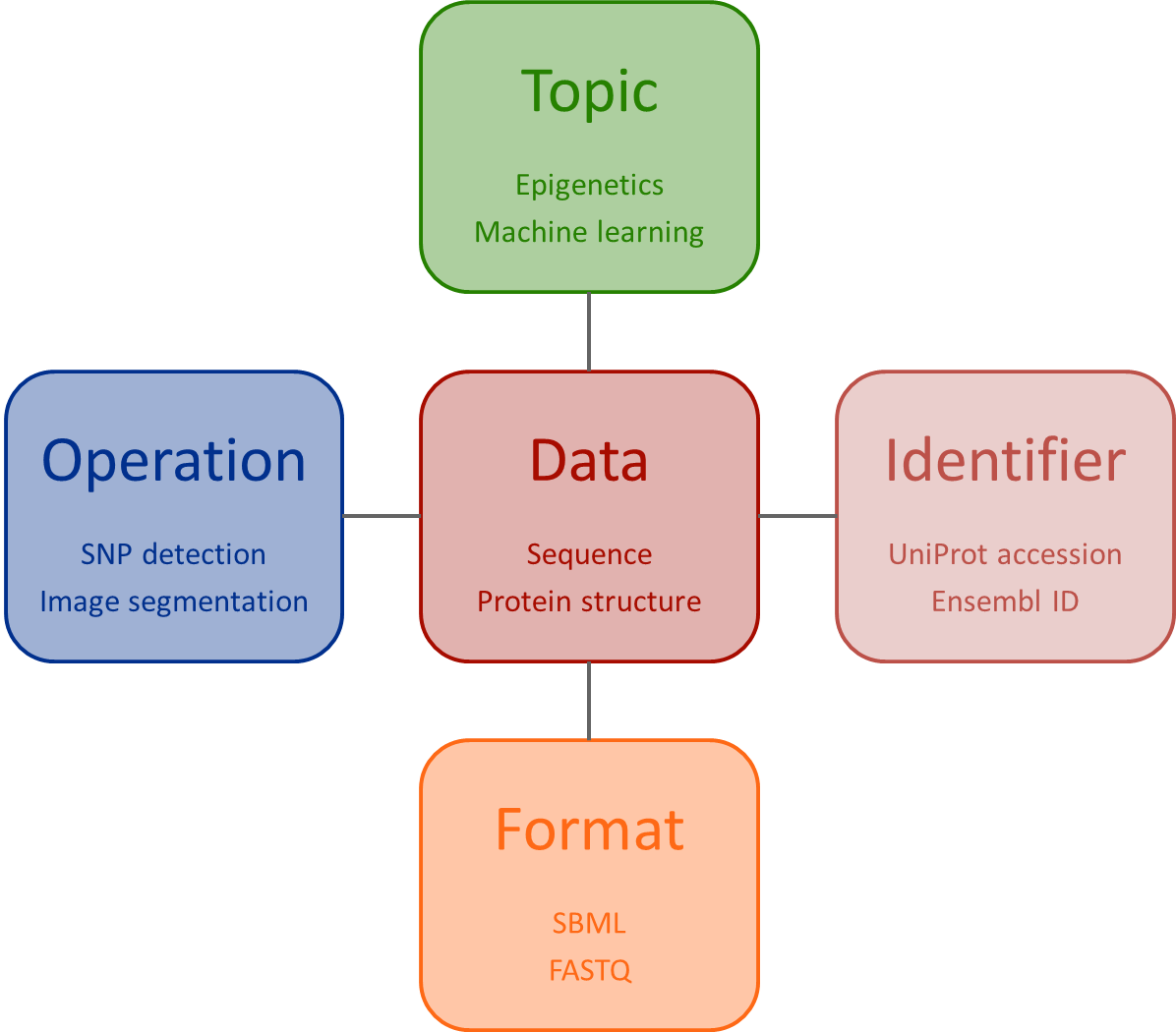
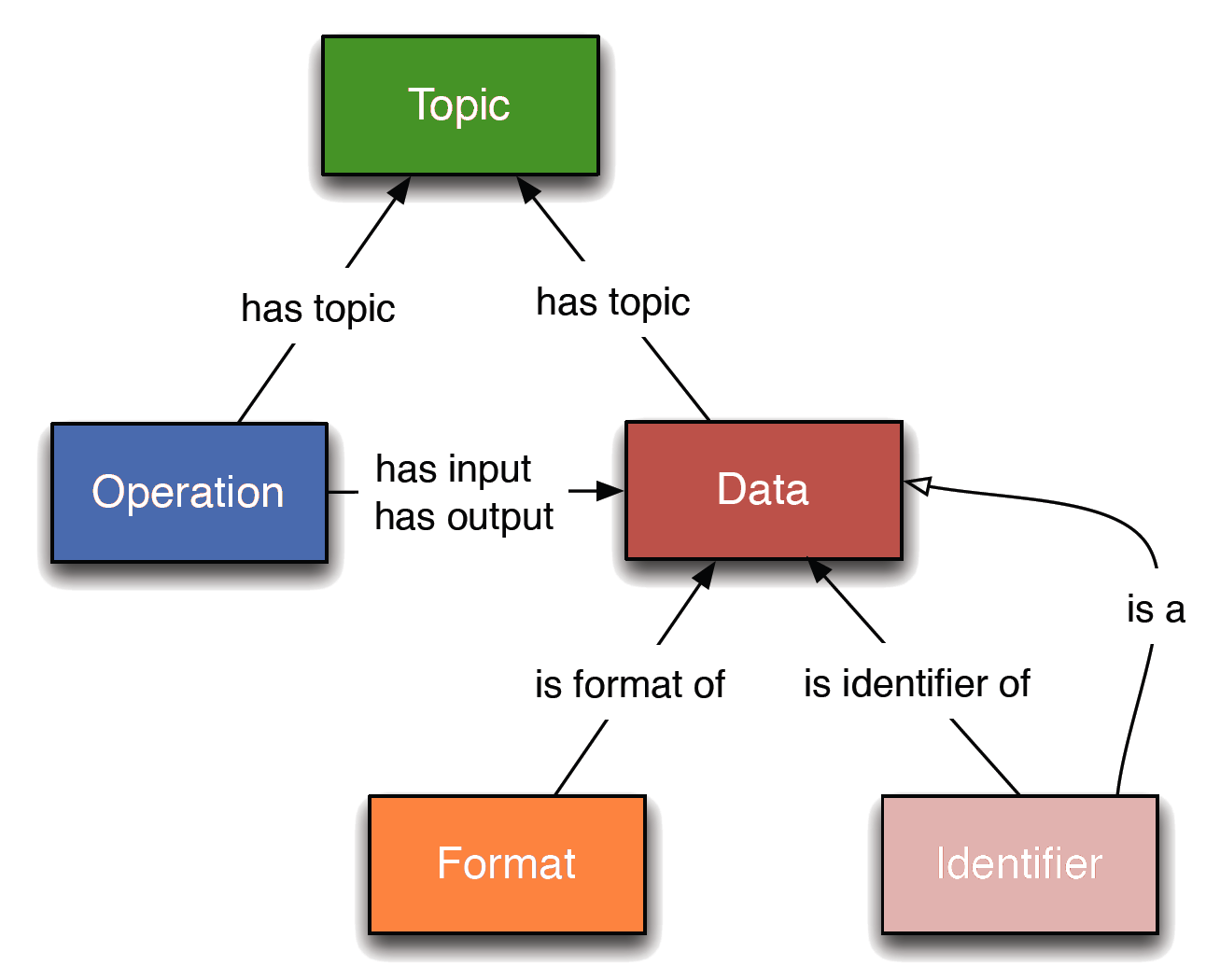
EDAM is a domain ontology of data analysis and data management in bio- and other sciences, and science-based applications. It comprises concepts related to analysis, modelling, optimisation, and data life-cycle. Targetting usability by diverse users, the structure of EDAM is relatively simple, divided into 4 main sections: Topic, Operation, Data (incl. Identifier), and Format.
EDAM is particularly suitable for semantic annotations and categorisation of diverse resources related to data analysis and management: e.g. tools, workflows, learning materials, or standards. EDAM is also useful in data management itself, for recording provenance metadata of processed data.
You can browse EDAM visually here:
- NCBO BioPortal (all-newest unstable version)
- OLS (latest stable version)
- EDAM Browser (new: all versions!)
- and other ontology browsers
Latest version
The very latest, unstable version:
Versioned releases:
- ..edamontology.org/EDAM_<x.y>.owl
EDAM comprises 5 main sections:
- Topic - A category denoting a rather broad domain or field of interest, of study, application, work, data, or technology.
- Operation - A function that processes a set of inputs and results in a set of outputs, or associates arguments (inputs) with values (outputs).
- Data - Information, represented in an information artefact (data record) that is 'understandable' by dedicated computational tools that can use the data as input or produce it as output.
- Data->Identifier - A text token, number or something else which identifies an entity, but which may not be persistent (stable) or unique (the same identifier may identify multiple things).
- Format - A defined way or layout of representing and structuring data in a computer file, blob, string, message, or elsewhere.
EDAM has 3 components:
- Concepts - All concepts have a preferred label (or 'term') and definition. Further, a concept may have simple relations (see below) to other EDAM concepts, as well other intrinsic properties, e.g. an identifier may have a regular expression defining its syntax, and formats have links to their documentation/specification.
- Hierarchy - Every concept (excluding top-level concepts) is related to one or more other concepts within the same branch by an is a (specialisation) relation. Hence EDAM has 4 primary hierarchies (for Data, Format, Operation, and Topic).
- Relations - Concepts are defined as sharing specific relationships (see figure below), reflecting well established or self-evident principles; these relationships are used primarily to define internal consistency of EDAM.
EDAM has been maturing steadily since its first creation ~2008. It is under active maintenance, developments are user-driven, for example by the Bio.tools registry, Galaxy, and other key infrastructure services. Future versions of EDAM will not depart fundamentally from the current structure (sections and relations). The development of EDAM can be followed on GitHub.
For ways to contribute, please see the documentation.
Our core priority is to be responsive to users of EDAM. Further, to ensure essential EDAM maintenance and development remain on a sustainable footing, including:
- Content review and refactoring to ensure structural and semantic simplicity ensuring high usability
- Community build-up and development including more formal, but agile, governance and maintenance models and mechanisms
- Agile and responsive development of content in close collaboration with end-users and serving concrete use-cases
- Technical refactoring to minimise the cost of routine housekeeping and content development
- Implementation of tooling for routine maintenance to serve the needs of end-users, e.g. harvesting change requests and mappings between concepts
EDAM strives to uphold a few founding principles including:
- Quality - an ontology that is moderated
- Openness - development by the open community
- Relevance - prioritising use-case-driven development towards comprehensive but practical coverage
- Practicality - practical utility is valued over ontological “strictness” or any metaphysical doctrine
- Clear scope - respecting the scope of other complementary, well-developed ontologies and open linked data efforts
- Familiarity - including only concepts that are well established; familiar are prevalent and jargon is discouraged
- Usability - conceptual hierarchy with sufficient richness but only necessary complexity
- Maintainability - development must be efficient and sustainably up to date in the long term
EDAM is working towards implementing these principles fully and is open to suggestions.
Scientists - professional and "citizen" - handle an increasingly large and diverse set of tools and data. Meanwhile, researchers demand ever more powerful and convenient means to organise, find, understand, compare, select, use and connect the available resources. These tasks often rely on consistent, machine-understandable descriptions of the underlying components, but these have been generally lacking in ad hoc resource descriptions. The urgent need - filled by EDAM - is for an ontology that unifies semantically the bioinformatics concepts in common use, provides the curator with a comprehensive controlled vocabulary that is broadly applicable, and supports new and powerful search, browse and query functions.
EDAM is suitable for large-scale semantic annotations and categorisation of diverse bioinformatics resources, including:
- Web APIs including RESTful APIs and SOAP/WSDL Web Services
- Application software
- Tool collections and packages
- Workflows / pipelines
- Databases
- XML Schemata and data objects
- Data syntax and file formats
- Web portals and pages
- Resource catalogues
- Training materials
- Courses, tutorials, and other events
- Documents, such as scientific publications
EDAM is suitable for diverse applications beyond annotation, for example within workbenches and workflow-management systems, software distributions, and resource registries.
An up-to-date description of EDAM is available in the following extended abstract and poster. If you use, or refer to EDAM or its part, please cite:
Melissa Black, Lucie Lamothe, Hager Eldakroury, Mads Kierkegaard, Ankita Priya, Anne Machinda, Uttam Singh Khanduja, Drashti Patoliya, Rashika Rathi, Tawah Peggy Che Nico, Gloria Umutesi, Claudia Blankenburg, Anita Op, Precious Chieke, Omodolapo Babatunde, Steve Laurie, Steffen Neumann, Veit Schwämmle, Ivan Kuzmin, Chris Hunter, Jonathan Karr, Jon Ison, Alban Gaignard, Bryan Brancotte, Hervé Ménager, Matúš Kalaš (2022). EDAM: the bioscientific data analysis ontology (update 2021) [version 1; not peer reviewed]. F1000Research, 11 (ISCB Comm J): 1. Poster. DOI: 10.7490/f1000research.1118900.1 Open access
The "source code" and releases of EDAM have DOIs:
- DOI representing all stable versions, resolving to the latest: 10.5281/zenodo.822690
- DOI of the latest stable version: https://zenodo.org/badge/latestdoi/20960594
The newest posters, available as PDF:
- EDAM (2022)
- EDAM Bioimaging (2020)
- EDAM Browser (2021)