diff --git a/.flake8 b/.flake8
index 2b138177..ada24518 100644
--- a/.flake8
+++ b/.flake8
@@ -1,6 +1,5 @@
[flake8]
ignore = D203, W503
-select = Q0
exclude =
# No need to traverse our git directory
.git,
@@ -8,4 +7,4 @@ exclude =
__pycache__,
max-complexity = 10
max-line-length = 100
-avoid-escape = True
\ No newline at end of file
+avoid-escape = True
diff --git a/.github/workflows/format_and_lint.yml b/.github/workflows/format_and_lint.yml
deleted file mode 100644
index 42dde253..00000000
--- a/.github/workflows/format_and_lint.yml
+++ /dev/null
@@ -1,37 +0,0 @@
-name: Python package
-
-on: [push, pull_request]
-
-jobs:
- build:
-
- runs-on: ubuntu-latest
- strategy:
- matrix:
-# python-version: ["3.8", "3.9", "3.10"]
- python-version: ["3.10"]
-
- steps:
- - uses: actions/checkout@v3
- - name: Set up Python ${{ matrix.python-version }}
- uses: actions/setup-python@v3
- with:
- python-version: ${{ matrix.python-version }}
- - name: Install dependencies
- run: |
- curl -sSL https://install.python-poetry.org | python3 - --version 1.4.0
- $HOME/.local/bin/poetry install
- - name: Check input line sorting with isort
- run: |
- source $(poetry env info --path)/bin/activate
- isort -c .
-
- - name: Check standarized formatting with yapf
- run: |
- source $(poetry env info --path)/bin/activate
- yapf -r --diff .
-
- - name: Lint with flake8
- run: |
- source $(poetry env info --path)/bin/activate
- flake8 . --count --show-source --statistics
diff --git a/.github/workflows/precommit_autoupdate.yaml b/.github/workflows/precommit_autoupdate.yaml
new file mode 100644
index 00000000..f67fd611
--- /dev/null
+++ b/.github/workflows/precommit_autoupdate.yaml
@@ -0,0 +1,40 @@
+# Adapted from https://github.com/browniebroke/browniebroke.com, Copyright (c) 2017 Bruno Allaname, available under the
+# MIT license
+
+on:
+ schedule:
+ - cron: '0 0 * * *'
+
+jobs:
+ autoupdate-precommit:
+ runs-on: ubuntu-latest
+ strategy:
+ matrix:
+# python-version: ["3.7", "3.8", "3.9", "3.10"]
+ python-version: ["3.10"]
+
+ steps:
+ - uses: actions/checkout@v3
+
+ - name: Set up Python
+ uses: actions/setup-python@v3
+ with:
+ python-version: ${{ matrix.python-version }}
+
+ - name: Install pre-commit
+ run: pip install pre-commit
+
+ - name: Run pre-commit autoupdate
+ run: pre-commit autoupdate
+
+ - name: Create Pull Request
+ uses: peter-evans/create-pull-request@v5
+ with:
+ token: ${{ secrets.CPR_GITHUB_TOKEN }}
+ branch: update/pre-commit-autoupdate
+ title: Auto-update pre-commit hooks
+ commit-message: Auto-update pre-commit hooks
+ body: |
+ Update versions of tools in pre-commit
+ configs to latest version
+ labels: dependencies
diff --git a/.github/workflows/run_precommit.yaml b/.github/workflows/run_precommit.yaml
new file mode 100644
index 00000000..f417e599
--- /dev/null
+++ b/.github/workflows/run_precommit.yaml
@@ -0,0 +1,29 @@
+name: Python package
+
+on: [push, pull_request]
+
+jobs:
+ run-precommit:
+ runs-on: ubuntu-latest
+ strategy:
+ matrix:
+# python-version: ["3.8", "3.9", "3.10"]
+ python-version: ["3.10"]
+
+ steps:
+ - uses: actions/checkout@v3
+ - name: Set up Python ${{ matrix.python-version }}
+ uses: actions/setup-python@v3
+ with:
+ python-version: ${{ matrix.python-version }}
+
+ - name: Install pre-commit
+ run: |
+ pip install pre-commit
+
+ - name: Run pre-commit autoupdate
+ run: pre-commit autoupdate
+
+ - name: Run pre-commit hooks with hook-stage "manual" (=checks only)
+ run: |
+ pre-commit run --hook-stage manual --all-files
diff --git a/.github/workflows/run_tests.yaml b/.github/workflows/run_tests.yaml
index a7ebf898..78a0e7d0 100644
--- a/.github/workflows/run_tests.yaml
+++ b/.github/workflows/run_tests.yaml
@@ -3,8 +3,7 @@ name: Python package
on: [push, pull_request]
jobs:
- build:
-
+ run-tests:
runs-on: ubuntu-latest
strategy:
matrix:
diff --git a/.github/workflows/todo_to_issue.yml b/.github/workflows/todo_to_issue.yaml
similarity index 70%
rename from .github/workflows/todo_to_issue.yml
rename to .github/workflows/todo_to_issue.yaml
index a694947e..1204c357 100644
--- a/.github/workflows/todo_to_issue.yml
+++ b/.github/workflows/todo_to_issue.yaml
@@ -1,9 +1,9 @@
name: "Run TODO to Issue"
on: ["push"]
jobs:
- build:
+ todo-to-issue:
runs-on: "ubuntu-latest"
steps:
- uses: "actions/checkout@v3"
- name: "TODO to Issue"
- uses: "alstr/todo-to-issue-action@v4"
\ No newline at end of file
+ uses: "alstr/todo-to-issue-action@v4"
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 00000000..09a4e3cc
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,47 @@
+repos:
+ - repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v4.4.0
+ hooks:
+ - id: check-yaml
+ types: [ yaml ]
+ - id: check-toml
+ types: [ toml ]
+ - id: check-merge-conflict
+ - id: check-ast
+ types: [ python ]
+ - repo: https://github.com/python-poetry/poetry
+ rev: 1.6.0
+ hooks:
+ - id: poetry-check
+ - repo: https://github.com/google/yapf
+ rev: v0.40.2
+ hooks:
+ - id: yapf
+ name: "yapf"
+ additional_dependencies: [toml]
+ args: [ '--style', 'pyproject.toml', '--parallel', '--in-place' ]
+ stages: [ commit ]
+ types: [ python ]
+ - id: yapf
+ name: "yapf (check)"
+ additional_dependencies: [toml]
+ args: [ '--style', 'pyproject.toml', '--parallel', '--diff' ]
+ stages: [ manual ]
+ types: [ python ]
+ - repo: https://github.com/pycqa/isort
+ rev: 5.12.0
+ hooks:
+ - id: isort
+ name: isort
+ stages: [ commit ]
+ types: [ python ]
+ - id: isort
+ name: isort (check)
+ args: [ '--check-only' ]
+ stages: [ manual ]
+ types: [ python ]
+ - repo: https://github.com/pycqa/flake8
+ rev: 6.1.0
+ hooks:
+ - id: flake8
+ additional_dependencies: [flake8-quotes==3.3.2]
diff --git a/CONTRIBUTING.md b/CONTRIBUTING.md
new file mode 100644
index 00000000..76590230
--- /dev/null
+++ b/CONTRIBUTING.md
@@ -0,0 +1,47 @@
+# Contributing to omnipy development
+
+## Development setup
+
+- Install Poetry:
+ - `curl -sSL https://install.python-poetry.org | python3 -`
+
+- Configure locally installed virtualenv (under `.venv`):
+ - `poetry config virtualenvs.in-project true`
+
+- Install dependencies:
+ - `poetry install --with dev --with docs`
+
+- Update all dependencies:
+ - `poetry update`
+
+- Update single dependency, e.g.:
+ - `poetry update prefect`
+
+- If a dependency is not updated to the latest version available on Pypi, you might need to clear
+ the pip cache of poetry:
+ - `poetry cache clear pypi --all`
+
+### For mypy support in PyCharm
+
+- In PyCharm, install "Mypy" plugin (not "Mypy (Official)")
+ - `which mypy` to get path to mypy binary
+ - In the PyCharm settings for the mypy plugin:
+ - Select the mypy binary
+ - Select `pyproject.toml` as the mypy config file
+
+### For automatic formatting and linting
+
+The setup for automatic formatting and linting is rather complex. The main alternative is to use
+black, which is easier to set up, but it does not have as many options and the main omnipy developer
+is opinionated against the default black setup. The yapf config is not fully
+defined.
+
+- To install git hooks that automagically format and lint before every commit:
+ - `pre-commit install`
+
+- To update pre-commit-managed dependencies to the latest repos' versions:
+ - `pre-commit autoupdate`
+
+- In PyCharm -> File Watchers:
+ - Click arrow icon pointing down and to the left
+ - Select `pycharm_file_watchers.xml`
diff --git a/INSTALL.md b/INSTALL.md
new file mode 100644
index 00000000..96e2b6e7
--- /dev/null
+++ b/INSTALL.md
@@ -0,0 +1,40 @@
+# Installation and usage of omnipy
+
+## Installation
+
+`pip install omnipy`
+
+
+## Run example scripts:
+ - Install `omnipy-examples`:
+ - `pip install omnipy-examples`
+ - Example script:
+ - `omnipy-examples isajson`
+ - For help on the command line interface:
+ - `omnipy-examples --help`
+ - For help on a particular example:
+ - `omnipy-examples isajson --help`
+
+### Output of flow runs
+
+The output will by default appear in the `data` directory, with a timestamp.
+
+ - It is recommended to install a file viewer that are capable of browsing tar.gz files.
+ For instance, the "File Expander" plugin in PyCharm is excellent for this.
+ - To unpack the compressed files of a run on the command line
+ (just make sure to replace the datetime string from this example):
+
+```
+for f in $(ls data/2023_02_03-12_51_51/*.tar.gz); do mkdir ${f%.tar.gz}; tar xfzv $f -C ${f%.tar.gz}; done
+```
+
+### Run with the Prefect engine
+
+Omnipy is integrated with the powerful [Prefect](https://prefect.io) data flow orchestration library.
+
+- To run an example using the `prefect` engine, e.g.:
+ - `omnipy-examples --engine prefect isajson`
+- After completion of some runs, you can check the flow logs and orchestration options in the Prefect UI:
+ - `prefect orion start`
+
+More info on Prefect configuration will come soon...
diff --git a/LICENSE b/LICENSE
index 261eeb9e..bc6ab255 100644
--- a/LICENSE
+++ b/LICENSE
@@ -186,7 +186,7 @@
same "printed page" as the copyright notice for easier
identification within third-party archives.
- Copyright [yyyy] [name of copyright owner]
+ Copyright 2023 Omnipy contributors
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
diff --git a/README.md b/README.md
index b6ad1183..3ba74c33 100644
--- a/README.md
+++ b/README.md
@@ -1,30 +1,44 @@
-

-Omnipy is the new name of the Python package formerly known as uniFAIR.
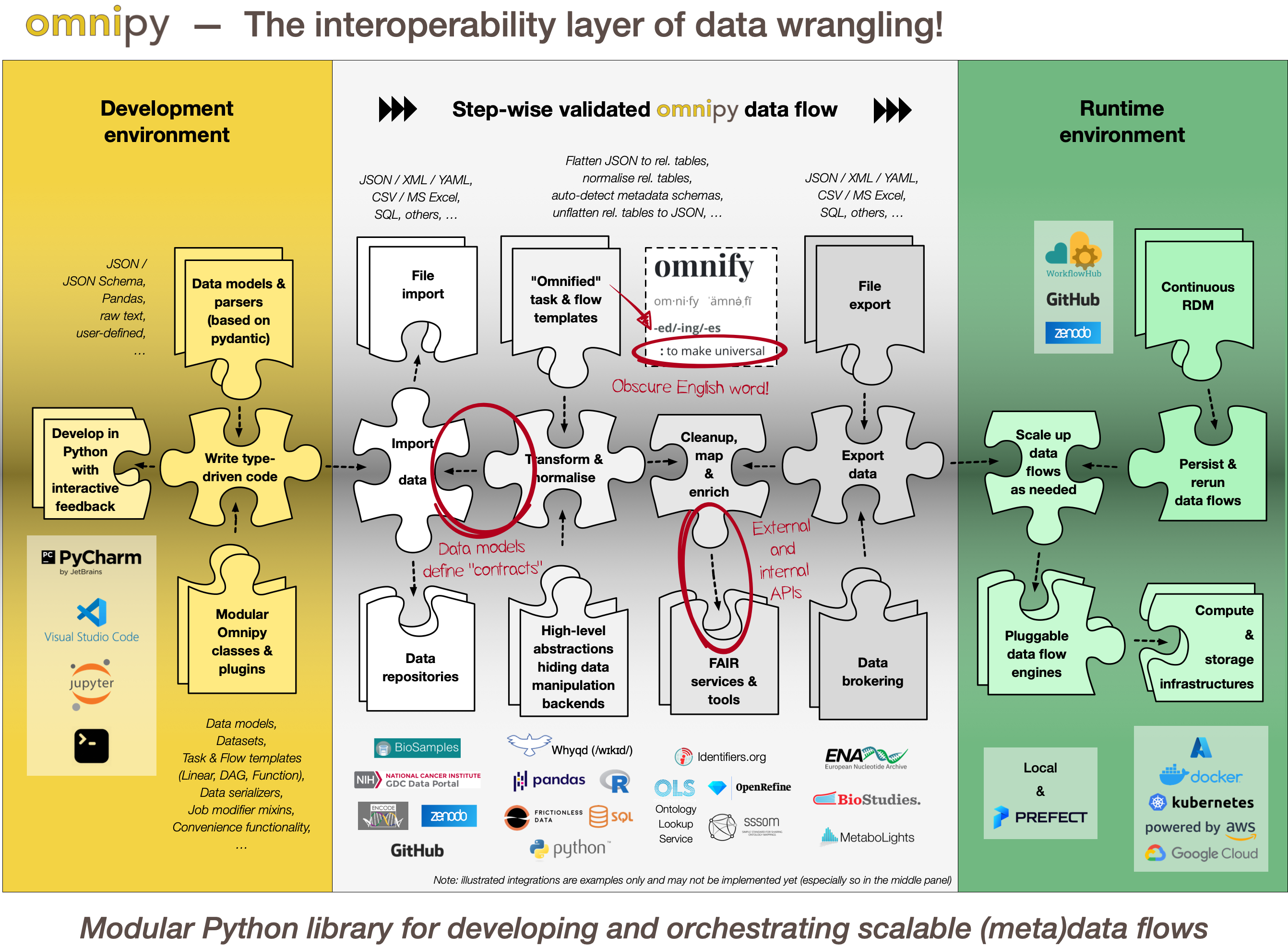
+Omnipy is a high level Python library for type-driven data wrangling and scalable workflow
+orchestration.
+
+
+
+# Updates
-We are very grateful to Dr. Jamin Chen, who gracefully transferred ownership of the (mostly unused) "omnipy" name in PyPI to us!
+- **Feb 3, 2023:** Documentation of the Omnipy API is still sparse. However, for examples of running
+ code, please check out the [omnipy-examples repo](https://github.com/fairtracks/omnipy_examples).
+- **Dec 22, 2022:** Omnipy is the new name of the Python package formerly known as uniFAIR.
+ _We are very grateful to Dr. Jamin Chen, who gracefully transferred ownership of the (mostly
+ unused) "omnipy" name in PyPI to us!__
---
+# Installation and use
-Update Feb 3, 2023: Documentation of the Omnipy API is still sparse. However, for running examples,
-please check out the [omnipy-examples repo](https://github.com/fairtracks/omnipy_examples) and its
-related [PYPI package](https://pypi.org/project/omnipy-examples/)!
+For basic information on installation and use of omnipy, read the [INSTALL.md](INSTALL.md)
+file.
-(NOTE: Read the section [Transformation on the FAIRtracks.net website](https://fairtracks.net/fair/#fair-07-transformation)
-for a more detailed and better formatted version of the following description!)
+# Contribute to omnipy development
+For basic information on how to set up a development environment to effectively contribute to
+the omnipy library, read the [CONTRIBUTING.md](CONTRIBUTING.md) file.
+
+# Overview of Omnipy
## Generic functionality
-Omnipy is designed primarily to simplify development and deployment of (meta)data transformation
-processes in the context of FAIRification and data brokering efforts. However, the functionality is
-very generic and can also be used to support research data (and metadata) transformations in a range
+_(NOTE: Read the
+section [Transformation on the FAIRtracks.net website](https://fairtracks.net/fair/#fair-07-transformation)
+for a more detailed and better formatted version of the following description!)_
+
+Omnipy is designed primarily to simplify development and deployment of (meta)data transformation
+processes in the context of FAIRification and data brokering efforts. However, the functionality is
+very generic and can also be used to support research data (and metadata) transformations in a range
of fields and contexts beyond life science, including day-to-day research scenarios:
-
+## Data wrangling in day-to-day research
-**Data wrangling in day-to-day research:** Researchers in life science and other data-centric fields
+Researchers in life science and other data-centric fields
often need to extract, manipulate and integrate data and/or metadata from different sources, such as
repositories, databases or flat files. Much research time is spent on trivial and not-so-trivial
details of such ["data wrangling"](https://en.wikipedia.org/wiki/Data_wrangling):
@@ -39,30 +53,35 @@ General software for data wrangling and analysis, such as [Pandas](https://panda
[R](https://www.r-project.org/) or [Frictionless](https://frictionlessdata.io/), are useful, but
researchers still regularly end up with hard-to-reuse scripts, often with manual steps.
-**Step-wise data model transformations:** With the Omnipy
-Python package, researchers can import (meta)data in almost any shape or form: _nested JSON; tabular
+## Step-wise data model transformations
+
+With the Omnipy Python package, researchers can import (meta)data in almost any shape or form:
+_nested JSON; tabular
(relational) data; binary streams; or other data structures_. Through a step-by-step process, data
is continuously parsed and reshaped according to a series of data model transformations.
-
-
+## "Parse, don't validate"
-**"Parse, don't validate":** Omnipy follows the principles of "Type-driven design" (read
-_Technical note #2: "Parse, don't validate"_ on the
-[FAIRtracks.net website](https://fairtracks.net/fair/#fair-07-transformation) for more info). It
+Omnipy follows the principles of "Type-driven design" (read
+_Technical note #2: "Parse, don't validate"_ on the
+[FAIRtracks.net website](https://fairtracks.net/fair/#fair-07-transformation) for more info). It
makes use of cutting-edge [Python type hints](https://peps.python.org/pep-0484/) and the popular
[pydantic](https://pydantic-docs.helpmanual.io/) package to "pour" data into precisely defined data
models that can range from very general (e.g. _"any kind of JSON data", "any kind of tabular data"_,
etc.) to very specific (e.g. _"follow the FAIRtracks JSON Schema for track files with the extra
restriction of only allowing BigBED files"_).
-**Data types as contracts:** Omnipy _tasks_ (single steps) or _flows_ (workflows) are defined as
+## Data types as contracts
+
+Omnipy _tasks_ (single steps) or _flows_ (workflows) are defined as
transformations from specific _input_ data models to specific _output_ data models.
[pydantic](https://pydantic-docs.helpmanual.io/)-based parsing guarantees that the input and output
data always follows the data models (i.e. data types). Thus, the data models defines "contracts"
that simplifies reuse of tasks and flows in a _mix-and-match_ fashion.
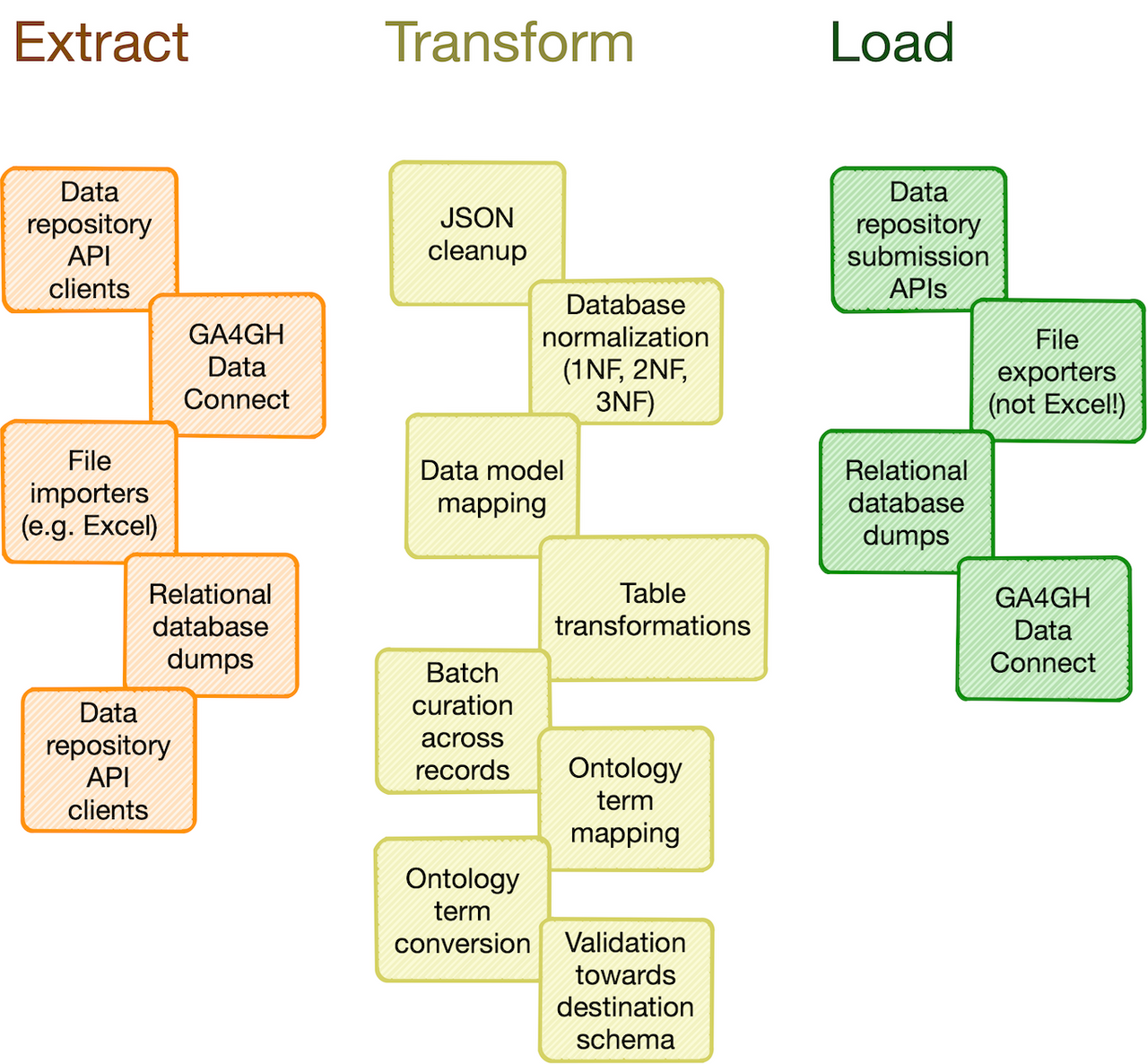
-**Catalog of common processing steps:** Omnipy is built from the ground up to be modular. We aim
+## Catalog of common processing steps
+
+Omnipy is built from the ground up to be modular. We aim
to provide a catalog of commonly useful functionality ranging from:
- data import from REST API endpoints, common flat file formats, database dumps, etc.
@@ -80,13 +99,17 @@ to transform metadata to follow the [FAIRtracks standard](/standards/#standards-
-**Refine and apply templates:** An Omnipy module typically consists of a set of generic _task_ and
+## Refine and apply templates
+
+An Omnipy module typically consists of a set of generic _task_ and
_flow templates_ with related data models, (de)serializers, and utility functions. The user can then
pick task and flow templates from this extensible, modular catalog, further refine them in the
context of a custom, use case-specific flow, and apply them to the desired compute engine to carry
out the transformations needed to wrangle data into the required shape.
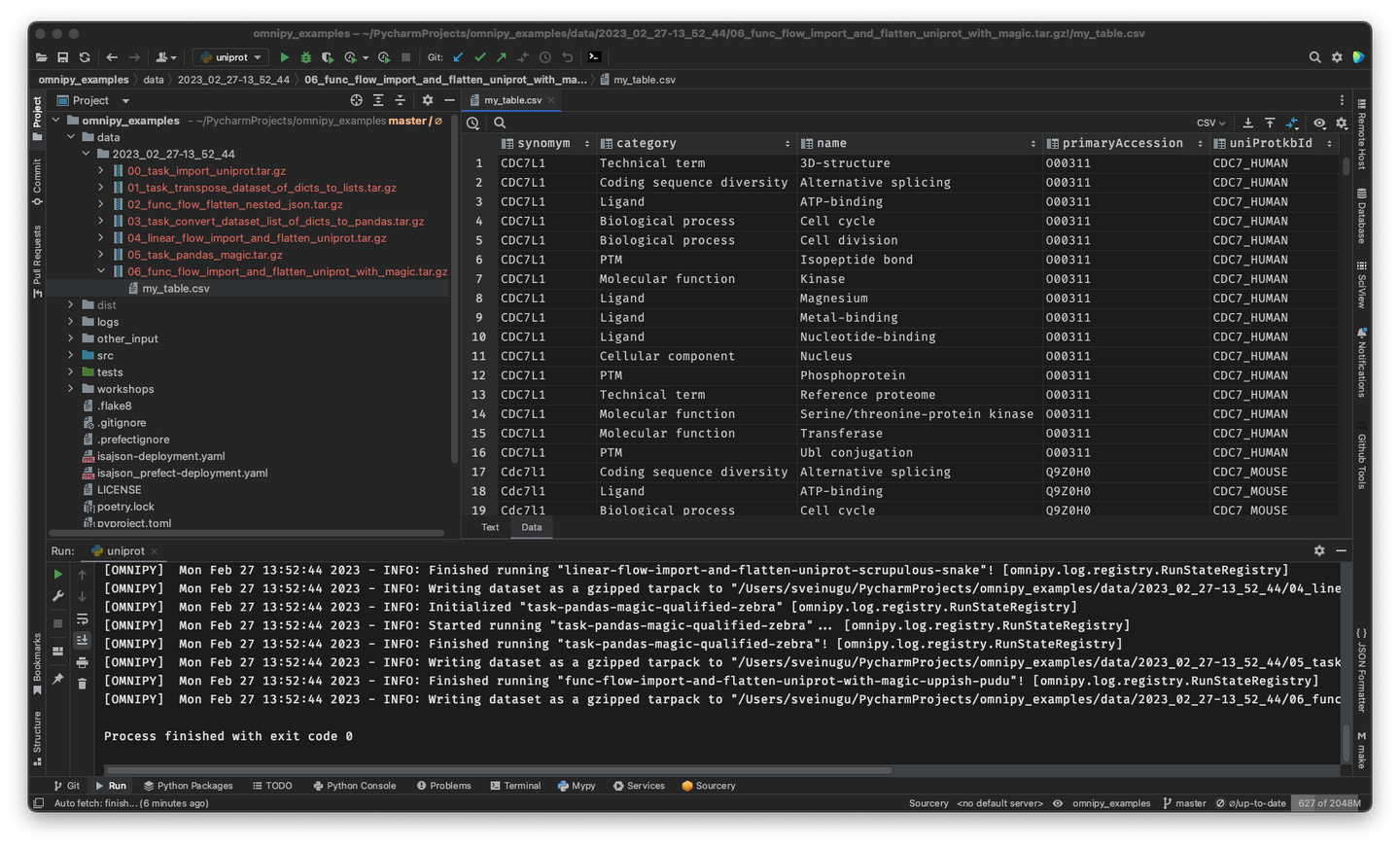
-**Rerun only when needed:** When piecing together a custom flow in Omnipy, the user has persistent
+## Rerun only when needed
+
+When piecing together a custom flow in Omnipy, the user has persistent
access to the state of the data at every step of the process. Persistent intermediate data allows
for caching of tasks based on the input data and parameters. Hence, if the input data and parameters
of a task does not change between runs, the task is not rerun. This is particularly useful for
@@ -94,7 +117,9 @@ importing from REST API endpoints, as a flow can be continuously rerun without t
server; data import will only carried out in the initial iteration or when the REST API signals that
the data has changed.
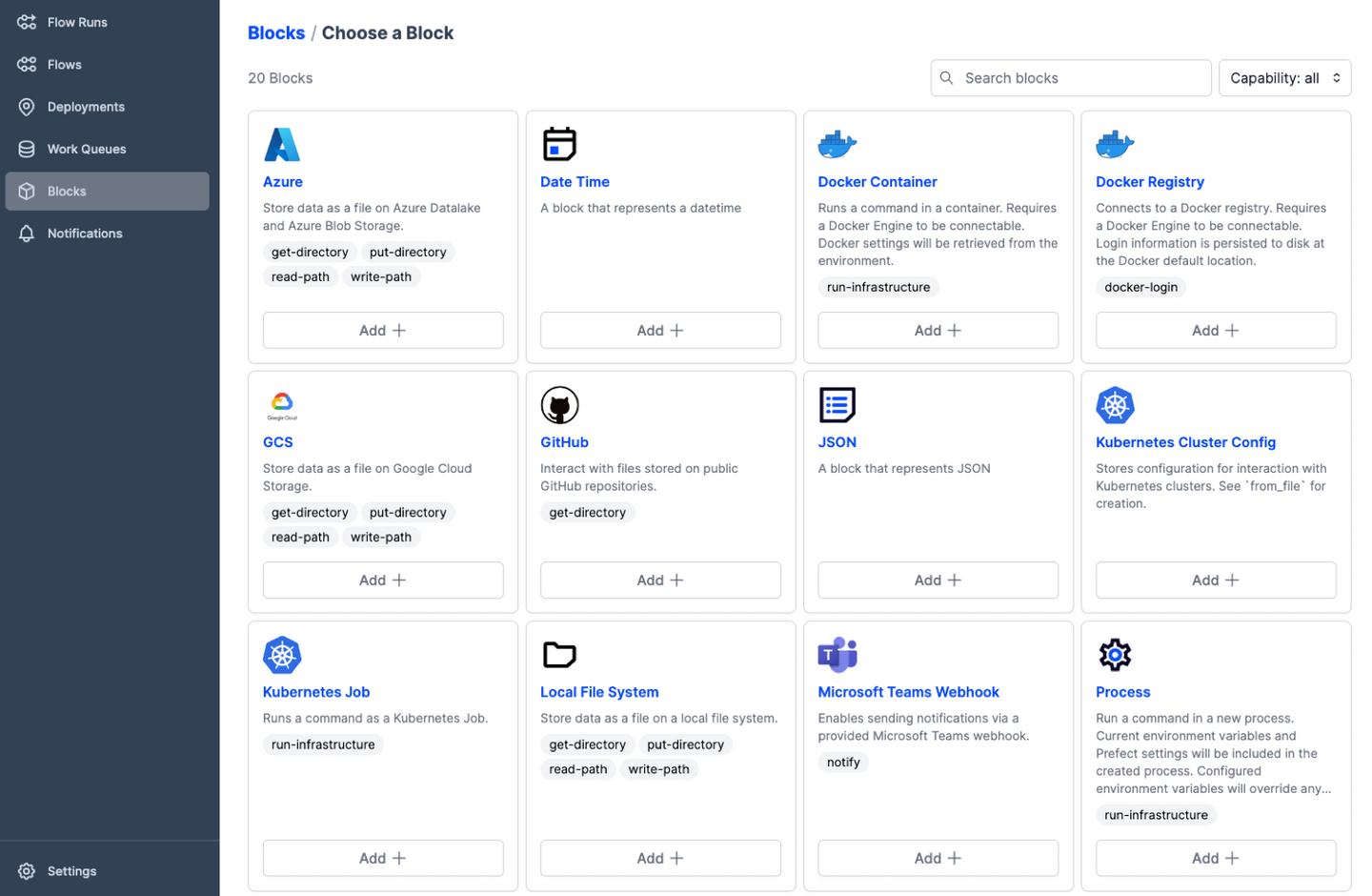
-**Scale up with external compute resources:** In the case of large datasets, the researcher can set
+## Scale up with external compute resources
+
+In the case of large datasets, the researcher can set
up a flow based on a representative sample of the full dataset, in a size that is suited for running
locally on, say, a laptop. Once the flow has produced the correct output on the sample data, the
operation can be seamlessly scaled up to the full dataset and sent off in
@@ -104,7 +129,9 @@ can be easily monitored using a web GUI.
-**Industry-standard ETL backbone:** Offloading of flows to external compute resources is provided by
+## Industry-standard ETL backbone
+
+Offloading of flows to external compute resources is provided by
the integration of Omnipy with a workflow engine based on the [Prefect](https://www.prefect.io/)
Python package. Prefect is an industry-leading platform for dataflow automation and orchestration
that brings a [series of powerful features](https://www.prefect.io/opensource/) to Omnipy:
@@ -121,225 +148,281 @@ that brings a [series of powerful features](https://www.prefect.io/opensource/)
-**Pluggable workflow engines:** It is also possible to integrate Omnipy with other workflow
+## Pluggable workflow engines
+
+It is also possible to integrate Omnipy with other workflow
backends by implementing new workflow engine plugins. This is relatively easy to do, as the core
architecture of Omnipy allows the user to easily switch the workflow engine at runtime. Omnipy
supports both traditional DAG-based and the more _avant garde_ code-based definition of flows. Two
workflow engines are currently supported: _local_ and _prefect_.
-## Scenarios
-As initial use cases, we will consider the following two scenarios:
+# Use cases (old notes)
+
+As initial use cases for the FAIRtracks project, we will consider the following two scenarios:
+
* Transforming ENCODE metadata into FAIRtracks format
* Transforming TCGA metadata into FAIRtracks format
## Nomenclature:
-* Omnipy is designed to work with content which could be classified both as data and metadata in their original context. For simplicity, we will refer to all such content as "data".
+
+* Omnipy is designed to work with content which could be classified both as data and metadata in
+ their original context. For simplicity, we will refer to all such content as "data".
## Overview of the proposed FAIRification process:
* ### Step 1: Import data from original source:
- * #### 1A: From API endpoints
- * _Input:_ API endpoint producing JSON data
- * _Output:_ JSON files (possibly with embedded JSON objects/lists [as strings])
- * _Description:_ General interface to support various API endpoints. Import all data by crawling API endpoints providing JSON content
- * _Generalizable:_ Partly (at least reuse of utility functions)
- * _Manual/automatic:_ Automatic
- * _Details:_
- * GDC/TCGA substeps (implemented as Step objects with file input/output)
- * 1A. Filtering step:
- * Input: parameters defining how to filter, e.g.:
- * For all endpoints (projects, cases, files, annotations), support:
- * Filter on list of IDs
- * Specific number of items
- * All
- * Example config:
- * projects: 2 items
- * cases: 2 items
- * files: all
- * annotations: all
- * Define standard configurations, e.g.:
- * Default: limited extraction (3 projects * 3 cases * 5 files? (+ annotations?))
- * All TCGA
- * List of projects
- * Hierarchical for loop through endpoints to create filter definitions
- * Output: Filter definitions as four files, e.g. as JSON,
- as they should be input to the filter parameter to the API:
- ```
- projects_filter.json:
- {
- "op": "in",
- "content": {
- "field": "project_id",
- "value": ['TCGA_ABCD', 'TCGA_BCDE']
- }
- }
-
- cases_filter.json:
- {
- "op": "in",
- "content": {
- "field": "case_id",
- "value": ['1234556', '234567', '3456789', '4567890']
- }
- }
-
- files_filter.json:
- {
- "op": "in",
- "content": {
- "field": "file_id",
- "value": ['1234556', '234567', '3456789', '4567890']
- }
- }
-
- annotations.json
- {
- "op": "in",
- "content": {
- "field": "annotation_id",
- "value": ['1234556', '234567', '3456789', '4567890']
- }
- }
- ```
-
- * 1B. Fetch and divide all fields step:
- * Input: None
- * Output: JSON files specifying all the fields of an endpoint fetched from the `mapping` API.
- The fields should be divided into chunks of a size that is small enough for the endpoints to
- handle. The JSON output should also specify the primary_key field, that needs to be added to
- all the API calls in order for the results to be joinable.
-
- Example JSON files:
- ```
- projects_fields.json:
- {
- "primary_key": "project_id",
- "fields_divided": [
- ["field_a", "field_b"],
- ["field_c.subfield_a", "field_c.subfield_b", "field_d"]
- ]
- }
-
- (...) # For all endpoints
- ```
- * 1C. Download from all endpoints according to the filters and the field divisions.
- If there is a limitation on the number of hits that the endpoint is able to return, divide into smaller
- API calls for a certain number of hits and concatenate the results. Make sure that proper waiting time
- (1 second?) is added between the calls (to not overload the endpoint).
- * 1D. Extract identifiers from nested objects (when present) and insert into parents objects
- * ENCODE:
- * Identify where to start (Cart? Experiment?)
- * To get all data for a table (double-check this): `https://www.encodeproject.org/experiments/@@listing?format=json&frame=object`
- * Download all tables directly.
- * #### 1b: From JSON files
- * _Input:_ JSON content as files
- * _Output:_ Pandas DataFrames (possibly with embedded JSON objects/lists)
- * _Description:_ Import data from files. Requires specific parsers to be implemented.
- * _Generalizable:_ Fully
- * _Manual/automatic:_ Automatic
- * #### 1c: From non-JSON files
- * _Input:_ File content in some supported format (e.g. GSuite)
- * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- * _Description:_ Import data from files. Requires specific parsers to be implemented.
- * _Generalizable:_ Partly (generating reference metadata might be tricky)
- * _Manual/automatic:_ Automatic
- * #### 1d: From database
- * _Input:_ Direct access to relational database
- * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- * _Description:_ Import data from database
- * _Generalizable:_ Fully
- * _Manual/automatic:_ Automatic
+ * #### 1A: From API endpoints
+ * _Input:_ API endpoint producing JSON data
+ * _Output:_ JSON files (possibly with embedded JSON objects/lists [as strings])
+ * _Description:_ General interface to support various API endpoints. Import all data by
+ crawling API endpoints providing JSON content
+ * _Generalizable:_ Partly (at least reuse of utility functions)
+ * _Manual/automatic:_ Automatic
+ * _Details:_
+ * GDC/TCGA substeps (implemented as Step objects with file input/output)
+ * 1A. Filtering step:
+ * Input: parameters defining how to filter, e.g.:
+ * For all endpoints (projects, cases, files, annotations), support:
+ * Filter on list of IDs
+ * Specific number of items
+ * All
+ * Example config:
+ * projects: 2 items
+ * cases: 2 items
+ * files: all
+ * annotations: all
+ * Define standard configurations, e.g.:
+ * Default: limited extraction (3 projects * 3 cases * 5 files? (+
+ annotations?))
+ * All TCGA
+ * List of projects
+ * Hierarchical for loop through endpoints to create filter definitions
+ * Output: Filter definitions as four files, e.g. as JSON,
+ as they should be input to the filter parameter to the API:
+ ```
+ projects_filter.json:
+ {
+ "op": "in",
+ "content": {
+ "field": "project_id",
+ "value": ['TCGA_ABCD', 'TCGA_BCDE']
+ }
+ }
+
+ cases_filter.json:
+ {
+ "op": "in",
+ "content": {
+ "field": "case_id",
+ "value": ['1234556', '234567', '3456789', '4567890']
+ }
+ }
+
+ files_filter.json:
+ {
+ "op": "in",
+ "content": {
+ "field": "file_id",
+ "value": ['1234556', '234567', '3456789', '4567890']
+ }
+ }
+
+ annotations.json
+ {
+ "op": "in",
+ "content": {
+ "field": "annotation_id",
+ "value": ['1234556', '234567', '3456789', '4567890']
+ }
+ }
+ ```
+
+ * 1B. Fetch and divide all fields step:
+ * Input: None
+ * Output: JSON files specifying all the fields of an endpoint fetched from
+ the `mapping` API.
+ The fields should be divided into chunks of a size that is small enough for
+ the endpoints to
+ handle. The JSON output should also specify the primary_key field, that needs
+ to be added to
+ all the API calls in order for the results to be joinable.
+
+ Example JSON files:
+ ```
+ projects_fields.json:
+ {
+ "primary_key": "project_id",
+ "fields_divided": [
+ ["field_a", "field_b"],
+ ["field_c.subfield_a", "field_c.subfield_b", "field_d"]
+ ]
+ }
+
+ (...) # For all endpoints
+ ```
+ * 1C. Download from all endpoints according to the filters and the field divisions.
+ If there is a limitation on the number of hits that the endpoint is able to
+ return, divide into smaller
+ API calls for a certain number of hits and concatenate the results. Make sure that
+ proper waiting time
+ (1 second?) is added between the calls (to not overload the endpoint).
+ * 1D. Extract identifiers from nested objects (when present) and insert into parents
+ objects
+ * ENCODE:
+ * Identify where to start (Cart? Experiment?)
+ * To get all data for a table (double-check
+ this): `https://www.encodeproject.org/experiments/@@listing?format=json&frame=object`
+ * Download all tables directly.
+ * #### 1b: From JSON files
+ * _Input:_ JSON content as files
+ * _Output:_ Pandas DataFrames (possibly with embedded JSON objects/lists)
+ * _Description:_ Import data from files. Requires specific parsers to be implemented.
+ * _Generalizable:_ Fully
+ * _Manual/automatic:_ Automatic
+ * #### 1c: From non-JSON files
+ * _Input:_ File content in some supported format (e.g. GSuite)
+ * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) +
+ reference metadata
+ * _Description:_ Import data from files. Requires specific parsers to be implemented.
+ * _Generalizable:_ Partly (generating reference metadata might be tricky)
+ * _Manual/automatic:_ Automatic
+ * #### 1d: From database
+ * _Input:_ Direct access to relational database
+ * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) +
+ reference metadata
+ * _Description:_ Import data from database
+ * _Generalizable:_ Fully
+ * _Manual/automatic:_ Automatic
* ### Step 2: JSON cleanup
* _Input:_ Pandas DataFrames (possibly with embedded JSON objects/lists)
- * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
+ * _Output:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) +
+ reference metadata
* _Description:_ Replace embedded objects with identifiers (possibly as lists)
* _Generalizable:_ Partly (generating reference metadata might be tricky)
* _Manual/automatic:_ Depending on original input
* _Details:_
- * If there are embedded objects from other tables:
- * ENCODE update:
- * By using the `frame=object` parameter, we will not get any embedded objects from the APIs for the main tables. There is, however, some "auditing" fields that contain JSON objects. We can ignore these in the first iteration.
- * If the original table of the embedded objects can be retrieved directly from an API, replace such embedded objects with unique identifiers to the object in another table (maintaining a reference to the name of the table, if needed)
- * Record the reference metadata `(table_from, attr_from) -> (table_to, attr_to)` for joins:
- * Example: `(table: "experiment", column: "replicates") -> (table: "replicate", column: "@id")`
- * If the original table of the embedded objects are not directly available from an API, one needs to fill out the other table with the content that is embedded in the current object, creating the table if needed.
- * For all fields with identifiers that reference other tables:
- * Record the reference metadata `(table_from, attr_from) -> (table_to, attr_to)` for joins.
- * If the field contains a list of identifiers
- * Convert into Pandas Series
+ * If there are embedded objects from other tables:
+ * ENCODE update:
+ * By using the `frame=object` parameter, we will not get any embedded objects from
+ the APIs for the main tables. There is, however, some "auditing" fields that
+ contain JSON objects. We can ignore these in the first iteration.
+ * If the original table of the embedded objects can be retrieved directly from an API,
+ replace such embedded objects with unique identifiers to the object in another table (
+ maintaining a reference to the name of the table, if needed)
+ * Record the reference metadata `(table_from, attr_from) -> (table_to, attr_to)` for
+ joins:
+ *
+ Example: `(table: "experiment", column: "replicates") -> (table: "replicate", column: "@id")`
+ * If the original table of the embedded objects are not directly available from an API,
+ one needs to fill out the other table with the content that is embedded in the current
+ object, creating the table if needed.
+ * For all fields with identifiers that reference other tables:
+ * Record the reference metadata `(table_from, attr_from) -> (table_to, attr_to)` for
+ joins.
+ * If the field contains a list of identifiers
+ * Convert into Pandas Series
* ### Step 3: Create reference tables to satisfy 1NF
- * _Input:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- * _Output:_ Pandas DataFrames (original tables without reference column) [1NF] + reference tables + reference metadata
- * _Description:_ Move references into separate tables, transforming the tables in first normal form ([1NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_1NF))
+ * _Input:_ Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) +
+ reference metadata
+ * _Output:_ Pandas DataFrames (original tables without reference column) [1NF] + reference
+ tables + reference metadata
+ * _Description:_ Move references into separate tables, transforming the tables in first normal
+ form ([1NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_1NF))
* _Generalizable:_ Fully
* _Manual/automatic:_ Automatic
* _Details:_
- * For each reference pair:
- * Create a reference table
- * For each item in the "from"-reference column:
- * Add new rows in the reference table for each "to"-identifier, using the same "from"-identifier
- * Example: Table "experiment-replicate" with columns "experiment.@id", "replicate.@id"
- * Delete the complete column from the original table
+ * For each reference pair:
+ * Create a reference table
+ * For each item in the "from"-reference column:
+ * Add new rows in the reference table for each "to"-identifier, using the same "
+ from"-identifier
+ * Example: Table "experiment-replicate" with columns "experiment.@id", "
+ replicate.@id"
+ * Delete the complete column from the original table
* ### Step 4: Satisfy 2NF
* _Input:_ Pandas DataFrames (original tables without reference column) [1NF] + reference tables
- * _Output:_ Pandas DataFrames (original tables without reference column) [2NF] + reference tables
- * _Description:_ Automatic transformation of original tables into second normal form ([2NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_2NF)):
+ * _Output:_ Pandas DataFrames (original tables without reference column) [2NF] + reference
+ tables
+ * _Description:_ Automatic transformation of original tables into second normal
+ form ([2NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_2NF)):
* _Generalizable:_ Fully (if not, we skip it)
* _Manual/automatic:_ Automatic
* _Details:_
- * Use existing library.
+ * Use existing library.
* ### Step 5: Satisfy 3NF
* _Input:_ Pandas DataFrames (original tables without reference column) [2NF] + reference tables
- * _Output:_ Pandas DataFrames (original tables without reference column) [3NF] + reference tables
- * _Description:_ Automatic transformation of original tables into third normal form ([3NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_3NF)):
+ * _Output:_ Pandas DataFrames (original tables without reference column) [3NF] + reference
+ tables
+ * _Description:_ Automatic transformation of original tables into third normal
+ form ([3NF](https://en.wikipedia.org/wiki/Database_normalization#Satisfying_3NF)):
* _Generalizable:_ Fully (if not, we skip it)
* _Manual/automatic:_ Automatic
* _Details:_
- * Use existing library.
+ * Use existing library.
* ### Step 6: Create model map
- * _Input:_ Pandas DataFrames (original tables without reference column) [Any NF] + reference tables + FAIRtracks JSON schemas
- * _Output:_ Model map [some data structure (to be defined) mapping FAIRtracks objects and attributes to tables/columns in the original data]
- * _Description:_ Manual mapping of FAIRtracks objects and attributes to corresponding tables and columns in the original data.
+ * _Input:_ Pandas DataFrames (original tables without reference column) [Any NF] + reference
+ tables + FAIRtracks JSON schemas
+ * _Output:_ Model
+ map [some data structure (to be defined) mapping FAIRtracks objects and attributes to tables/columns in the original data]
+ * _Description:_ Manual mapping of FAIRtracks objects and attributes to corresponding tables and
+ columns in the original data.
* _Generalizable:_ Fully
* _Manual/automatic:_ Manual
* Details:
- * For each FAIRtracks object:
- * Define a start table in the original data
- * For each FAIRtracks attribute:
- * Manually find the path (or paths) to the original table/column that this maps to
- * _Example:_ `Experiments:organism (FAIRtracks) -> Experiments.Biosamples.Organism.scientific_name`
+ * For each FAIRtracks object:
+ * Define a start table in the original data
+ * For each FAIRtracks attribute:
+ * Manually find the path (or paths) to the original table/column that this maps to
+ *
+ _Example:_ `Experiments:organism (FAIRtracks) -> Experiments.Biosamples.Organism.scientific_name`
* ### Step 7: Apply model map to generate initial FAIRtracks tables
- * _Input:_ Pandas DataFrames (original tables without reference column) [Any NF] + reference tables + Model map
- * _Output:_ Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes)
- * Example: `Experiment.target_from_origcolumn1` and `Experimentl.target_from_origcolumn2` contain content from two different attributes from the original data that both corresponds to `Experiment.target`
- * _Description:_ Generate initial FAIRtracks tables by applying the model map, mapping FAIRtracks attributes with one or more attributes (columns) in the original table.
- * _Generalizable:_ Fully
- * _Manual/automatic:_ Automatic
- * _Details:_
- * For every FAIRtracks object:
- * Create a new pandas DataFrame
- * For every FAIRtracks attribute:
- * From the model map, get the path to the corresponding original table/column, or a list of such paths in case of multimapping
- * For each path:
- * Automatically join tables to get primary keys and attribute value in the same table:
- * _Example:_ `experiment-biosample JOIN biosample-organism JOIN organism` will create mapping table with two columns: `Experiments.local_id` and `Organism.scientific_name`
- * Add column to FAIRtracks DataFrame
- * In case of multimodeling, record the relation between FAIRtracks attribute and corresponding multimapped attributes, e.g. by generating unique attribute names for each path, such as `Experiment.target_from_origcolumn1` and `Experiment.target_from_origcolumn2`, which one can derive directly from the model map.
+ * _Input:_ Pandas DataFrames (original tables without reference column) [Any NF] + reference
+ tables + Model map
+ * _Output:_ Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes)
+ * Example: `Experiment.target_from_origcolumn1` and `Experimentl.target_from_origcolumn2`
+ contain content from two different attributes from the original data that both corresponds
+ to `Experiment.target`
+ * _Description:_ Generate initial FAIRtracks tables by applying the model map, mapping
+ FAIRtracks attributes with one or more attributes (columns) in the original table.
+ * _Generalizable:_ Fully
+ * _Manual/automatic:_ Automatic
+ * _Details:_
+ * For every FAIRtracks object:
+ * Create a new pandas DataFrame
+ * For every FAIRtracks attribute:
+ * From the model map, get the path to the corresponding original table/column, or a
+ list of such paths in case of multimapping
+ * For each path:
+ * Automatically join tables to get primary keys and attribute value in the same
+ table:
+ * _Example:_ `experiment-biosample JOIN biosample-organism JOIN organism`
+ will create mapping table with two columns: `Experiments.local_id`
+ and `Organism.scientific_name`
+ * Add column to FAIRtracks DataFrame
+ * In case of multimodeling, record the relation between FAIRtracks attribute and
+ corresponding multimapped attributes, e.g. by generating unique attribute
+ names for each path, such as `Experiment.target_from_origcolumn1`
+ and `Experiment.target_from_origcolumn2`, which one can derive directly from
+ the model map.
* ### Step 8: Harmonize multimapped attributes
- * _Input:_ Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes) + model map
- * _Output:_ Pandas DataFrames (initial FAIRtracks tables)
- * _Description:_ Harmonize multimapped attributes manually, or possibly by applying scripts
- * _Generalizable:_ Limited (mostly by reusing util functions)
- * _Manual/automatic:_ Mixed (possibly scriptable)
- * _Details:_
- * For all multimapped attributes:
- * Manually review values (in batch mode) and generate a single output value for each combination:
- * Hopefully Open Refine can be used for this. If so, one needs to implement data input/output mechanisms.
+ * _Input:_ Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes) +
+ model map
+ * _Output:_ Pandas DataFrames (initial FAIRtracks tables)
+ * _Description:_ Harmonize multimapped attributes manually, or possibly by applying scripts
+ * _Generalizable:_ Limited (mostly by reusing util functions)
+ * _Manual/automatic:_ Mixed (possibly scriptable)
+ * _Details:_
+ * For all multimapped attributes:
+ * Manually review values (in batch mode) and generate a single output value for each
+ combination:
+ * Hopefully Open Refine can be used for this. If so, one needs to implement data
+ input/output mechanisms.
* ### Further steps to be detailed:
- * For all FAIRtracks attributes with ontology terms: Convert terms using required ontologies
- * Other FAIRtracks specific value conversion
- * Manual batch correction of values (possibly with errors), probably using Open Refine
- * Validation of FAIRtracks document
+ * For all FAIRtracks attributes with ontology terms: Convert terms using required ontologies
+ * Other FAIRtracks specific value conversion
+ * Manual batch correction of values (possibly with errors), probably using Open Refine
+ * Validation of FAIRtracks document
-Suggestion: we will use Pandas DataFrames as the core data structure for tables, given that the library provides the required features (specifically Foreign key and Join capabilities)
+Suggestion: we will use Pandas DataFrames as the core data structure for tables, given that the
+library provides the required features (specifically Foreign key and Join capabilities)
diff --git a/docs/examples/dynamic_mixin_example.py b/docs/examples/dynamic_mixin_example.py
index efb0a3de..3324f54b 100644
--- a/docs/examples/dynamic_mixin_example.py
+++ b/docs/examples/dynamic_mixin_example.py
@@ -136,9 +136,8 @@ def instantiate_and_print_results(d_cls, d_obj_name, header):
print(f'{d_obj_name}.__class__.__bases__={d_base_classes}')
for i in range(len(d_base_classes)):
- print(
- f'{d_obj_name}.__class__.__bases__[{i}].__bases__={d_obj.__class__.__bases__[i].__bases__}'
- )
+ print(f'{d_obj_name}.__class__.__bases__[{i}].__bases__='
+ + d_obj.__class__.__bases__[i].__bases__)
print()
print(f'{d_obj_name} = {d_cls.__name__}()')
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 3df7f495..16f1785b 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -10,9 +10,11 @@
# https://www.sphinx-doc.org/en/master/usage/configuration.html#project-information
project = 'Omnipy'
-copyright = '2023, Sveinung Gundersen, Joshua Baskaran, Federico Bianchini, Jeanne Cheneby, Ahmed Ghanem, Pável Vázquez'
-author = 'Sveinung Gundersen, Joshua Baskaran, Federico Bianchini, Jeanne Cheneby, Ahmed Ghanem, Pável Vázquez'
-release = '0.10.4'
+copyright = '2023, Sveinung Gundersen, Joshua Baskaran, Federico Bianchini, Jeanne Cheneby, ' \
+ 'Ahmed Ghanem, Pável Vázquez'
+author = 'Sveinung Gundersen, Joshua Baskaran, Federico Bianchini, Jeanne Cheneby, Ahmed Ghanem, ' \
+ 'Pável Vázquez'
+release = '0.10.5'
# -- General configuration ---------------------------------------------------
# https://www.sphinx-doc.org/en/master/usage/configuration.html#general-configuration
diff --git a/pycharm-file-watchers.xml b/pycharm-file-watchers.xml
new file mode 100644
index 00000000..53fede9e
--- /dev/null
+++ b/pycharm-file-watchers.xml
@@ -0,0 +1,62 @@
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
\ No newline at end of file
diff --git a/pyproject.toml b/pyproject.toml
index 66651f47..c171f0a0 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -37,16 +37,13 @@ typing-inspect = "^0.8.0"
[tool.poetry.group.dev.dependencies]
deepdiff = "^6.2.1"
-flake8 = "^6.0.0"
-flake8-quotes = "^3.3.1"
-isort = "^5.10.1"
mypy = "^1.1.1"
pytest = "^7.1.0"
pytest-pycharm = "^0.7.0"
types-python-slugify = "^8.0.0.1"
-yapf = "^0.32.0"
types-requests = "^2.28.11.15"
pandas-stubs = "^1.5.3.230304"
+pre-commit = "^2.15.0"
[tool.poetry.group.docs.dependencies]
furo = "^2022.12.7"
diff --git a/src/omnipy/__init__.py b/src/omnipy/__init__.py
index 9d8747b2..e4a5c17e 100644
--- a/src/omnipy/__init__.py
+++ b/src/omnipy/__init__.py
@@ -1,18 +1,20 @@
-__version__ = '0.10.4'
+__version__ = '0.10.5'
import os
import sys
from typing import Optional
+from omnipy.hub.runtime import Runtime
+
ROOT_DIR = os.path.dirname(os.path.abspath(__file__))
-# TODO: The check disabling runtime for tests also trigger for tests that are run outside of Omnipy, breaking
-# tests on the user side. Find a better way to disable the global runtime object for Omnipy tests
+# TODO: The check disabling runtime for tests also trigger for tests that are run outside of Omnipy,
+# breaking tests on the user side.
+# Find a better way to disable the global runtime object for Omnipy tests
def _get_runtime() -> Optional['Runtime']:
if 'pytest' not in sys.modules:
- from omnipy.hub.runtime import Runtime
return Runtime()
else:
return None
diff --git a/src/omnipy/api/protocols/public/hub.py b/src/omnipy/api/protocols/public/hub.py
index 9ff45ee4..c4329eac 100644
--- a/src/omnipy/api/protocols/public/hub.py
+++ b/src/omnipy/api/protocols/public/hub.py
@@ -2,7 +2,7 @@
import logging
from logging.handlers import TimedRotatingFileHandler
-from typing import Any, Optional, Protocol
+from typing import Optional, Protocol
from omnipy.api.enums import EngineChoice
from omnipy.api.protocols.private.compute.job_creator import IsJobConfigHolder
diff --git a/src/omnipy/compute/mixins/mixin_types.py b/src/omnipy/compute/mixins/mixin_types.py
index db3763d0..62ee0f2a 100644
--- a/src/omnipy/compute/mixins/mixin_types.py
+++ b/src/omnipy/compute/mixins/mixin_types.py
@@ -1,4 +1,4 @@
-from typing import Protocol, Type, TypeVar, Union
+from typing import Protocol, TypeVar, Union
from omnipy.data.dataset import Dataset
from omnipy.data.model import Model
diff --git a/src/omnipy/compute/mixins/params.py b/src/omnipy/compute/mixins/params.py
index ac341204..b626afca 100644
--- a/src/omnipy/compute/mixins/params.py
+++ b/src/omnipy/compute/mixins/params.py
@@ -35,7 +35,7 @@ def param_key_map(self) -> MappingProxyType[str, str]:
def _call_job(self, *args: object, **kwargs: object) -> object:
self_as_name_job_base_mixin = cast(NameJobBaseMixin, self)
- self_signature_func_job_base_mixin = cast(SignatureFuncJobBaseMixin, self)
+ self_as_signature_func_job_base_mixin = cast(SignatureFuncJobBaseMixin, self)
mapped_fixed_params = self._param_key_mapper.delete_matching_keys(
self._fixed_params, inverse=True)
@@ -51,17 +51,17 @@ def _call_job(self, *args: object, **kwargs: object) -> object:
except TypeError as e:
if str(e).startswith('Incorrect job function arguments'):
- raise TypeError(
- f'Incorrect job function arguments for job "{self_as_name_job_base_mixin.name}"!\n'
- f'Job class name: {self.__class__.__name__}\n'
- f'Current parameter key map contents: {self.param_key_map}\n'
- f'Positional arguments: {repr_max_len(args)}\n'
- f'Keyword arguments: {repr_max_len(kwargs)}\n'
- f'Mapped fixed parameters: {repr_max_len(mapped_fixed_params)}\n'
- f'Mapped keyword arguments: {repr_max_len(mapped_kwargs)}\n'
- f'Call function signature parameters: '
- f'{[(str(p), p.kind) for p in self_signature_func_job_base_mixin.param_signatures.values()]}'
- ) from e
+ signature_values = self_as_signature_func_job_base_mixin.param_signatures.values()
+ raise TypeError(f'Incorrect job function arguments for job '
+ f'"{self_as_name_job_base_mixin.name}"!\n'
+ f'Job class name: {self.__class__.__name__}\n'
+ f'Current parameter key map contents: {self.param_key_map}\n'
+ f'Positional arguments: {repr_max_len(args)}\n'
+ f'Keyword arguments: {repr_max_len(kwargs)}\n'
+ f'Mapped fixed parameters: {repr_max_len(mapped_fixed_params)}\n'
+ f'Mapped keyword arguments: {repr_max_len(mapped_kwargs)}\n'
+ f'Call function signature parameters: '
+ f'{[(str(p), p.kind) for p in signature_values]}') from e
else:
raise
diff --git a/src/omnipy/compute/mixins/serialize.py b/src/omnipy/compute/mixins/serialize.py
index 173b66c9..a1f5bc8f 100644
--- a/src/omnipy/compute/mixins/serialize.py
+++ b/src/omnipy/compute/mixins/serialize.py
@@ -2,7 +2,7 @@
import os
from pathlib import Path
import tarfile
-from typing import cast, Optional, Type
+from typing import cast, Generator, Optional, Type
from omnipy.api.enums import ConfigPersistOutputsOptions as ConfigPersistOpts
from omnipy.api.enums import ConfigRestoreOutputsOptions as ConfigRestoreOpts
@@ -86,7 +86,7 @@ def restore_outputs(self) -> Optional[RestoreOutputsOptions]:
def will_persist_outputs(self) -> PersistOutputsOptions:
if not self._has_job_config or self._persist_outputs is not PersistOpts.FOLLOW_CONFIG:
return self._persist_outputs if self._persist_outputs is not None \
- else PersistOpts.DISABLED
+ else PersistOpts.DISABLED
else:
# TODO: Refactor using Flow and Task Mixins
from omnipy.compute.flow import FlowBase
@@ -98,8 +98,8 @@ def will_persist_outputs(self) -> PersistOutputsOptions:
return PersistOpts.ENABLED if isinstance(self, FlowBase) else PersistOpts.DISABLED
elif config_persist_opt == ConfigPersistOpts.ENABLE_FLOW_AND_TASK_OUTPUTS:
return PersistOpts.ENABLED \
- if any(isinstance(self, cls) for cls in (FlowBase, TaskBase)) \
- else PersistOpts.DISABLED
+ if any(isinstance(self, cls) for cls in (FlowBase, TaskBase)) \
+ else PersistOpts.DISABLED
else:
assert config_persist_opt == ConfigPersistOpts.DISABLED
return PersistOpts.DISABLED
@@ -108,7 +108,7 @@ def will_persist_outputs(self) -> PersistOutputsOptions:
def will_restore_outputs(self) -> RestoreOutputsOptions:
if not self._has_job_config or self._restore_outputs is not RestoreOpts.FOLLOW_CONFIG:
return self._restore_outputs if self._restore_outputs is not None \
- else RestoreOpts.DISABLED
+ else RestoreOpts.DISABLED
else:
config_restore_opt = self._job_config.restore_outputs
@@ -137,8 +137,8 @@ def _call_job(self, *args: object, **kwargs: object) -> object:
self._serialize_and_persist_outputs(results)
else:
self._log(
- f'Results of {self_as_name_job_base_mixin.unique_name} is not a Dataset and cannot '
- f'be automatically serialized and persisted!')
+ f'Results of {self_as_name_job_base_mixin.unique_name} is not a Dataset and '
+ f'cannot be automatically serialized and persisted!')
return results
@@ -152,7 +152,7 @@ def _serialize_and_persist_outputs(self, results: Dataset):
os.makedirs(output_path)
num_cur_files = len(os.listdir(output_path))
- job_name = self._create_job_name()
+ job_name = self._job_name()
file_path = output_path.joinpath(f'{num_cur_files:02}_{job_name}.tar.gz')
@@ -160,16 +160,16 @@ def _serialize_and_persist_outputs(self, results: Dataset):
self._serializer_registry.auto_detect_tar_file_serializer(results)
if serializer is None:
- self._log(
- f'Unable to find a serializer for results of job "{self_as_name_job_base_mixin.name}", '
- f'with data type "{type(results)}". Will abort persisting results...')
+ self._log('Unable to find a serializer for results of job '
+ f'"{self_as_name_job_base_mixin.name}", with data type "{type(results)}". '
+ f'Will abort persisting results...')
else:
self._log(f'Writing dataset as a gzipped tarpack to "{os.path.abspath(file_path)}"')
with open(file_path, 'wb') as tarfile:
tarfile.write(serializer.serialize(parsed_dataset))
- def _create_job_name(self):
+ def _job_name(self):
return '_'.join(self.unique_name.split('-')[:-2])
def _generate_datetime_str(self):
@@ -183,53 +183,60 @@ def _generate_datetime_str(self):
datetime_str = run_time.strftime('%Y_%m_%d-%H_%M_%S')
return datetime_str
- # TODO: Refactor
+ @staticmethod
+ def _all_job_output_file_paths_in_reverse_order_for_last_run(
+ persist_data_dir_path: Path, job_name: str) -> Generator[Path, None, None]:
+ sorted_date_dirs = list(sorted(os.listdir(persist_data_dir_path)))
+ if len(sorted_date_dirs) > 0:
+ last_dir = sorted_date_dirs[-1]
+ last_dir_path = persist_data_dir_path.joinpath(last_dir)
+ for job_output_name in reversed(sorted(os.listdir(last_dir_path))):
+ name_part_of_filename = job_output_name[3:-7]
+ if name_part_of_filename == job_name:
+ yield last_dir_path.joinpath(job_output_name)
+ else:
+ raise StopIteration
+
+ # TODO: Further refactor _deserialize_and_restore_outputs
def _deserialize_and_restore_outputs(self) -> Dataset:
- output_path = Path(self._job_config.persist_data_dir_path)
- if os.path.exists(output_path):
- sorted_date_dirs = list(sorted(os.listdir(output_path)))
- if len(sorted_date_dirs) > 0:
- last_dir = sorted_date_dirs[-1]
- last_dir_path = output_path.joinpath(last_dir)
- for job_output_name in reversed(sorted(os.listdir(last_dir_path))):
- name_part_of_filename = job_output_name[3:-7]
- if name_part_of_filename == self._create_job_name():
- tar_file_path = last_dir_path.joinpath(job_output_name)
- with tarfile.open(tar_file_path, 'r:gz') as tarfile_obj:
- file_suffixes = set(fn.split('.')[-1] for fn in tarfile_obj.getnames())
- if len(file_suffixes) != 1:
- self._log(f'Tar archive contains files with different or '
- f'no file suffixes: {file_suffixes}. Serializer '
- f'cannot be uniquely determined. Aborting '
- f'restore.')
+ persist_data_dir_path = Path(self._job_config.persist_data_dir_path)
+ if os.path.exists(persist_data_dir_path):
+ for tar_file_path in self._all_job_output_file_paths_in_reverse_order_for_last_run(
+ persist_data_dir_path, self._job_name()):
+ with tarfile.open(tar_file_path, 'r:gz') as tarfile_obj:
+ file_suffixes = set(fn.split('.')[-1] for fn in tarfile_obj.getnames())
+ if len(file_suffixes) != 1:
+ self._log(f'Tar archive contains files with different or '
+ f'no file suffixes: {file_suffixes}. Serializer '
+ f'cannot be uniquely determined. Aborting '
+ f'restore.')
+ else:
+ file_suffix = file_suffixes.pop()
+ serializers = self._serializer_registry.\
+ detect_tar_file_serializers_from_file_suffix(file_suffix)
+ if len(serializers) == 0:
+ self._log(f'No serializer for file suffix "{file_suffix}" can be'
+ f'determined. Aborting restore.')
+ else:
+ self._log(f'Reading dataset from a gzipped tarpack at'
+ f' "{os.path.abspath(tar_file_path)}"')
+
+ serializer = serializers[0]
+ with open(tar_file_path, 'rb') as tarfile_binary:
+ dataset = serializer.deserialize(tarfile_binary.read())
+ return_dataset_cls = cast(Type[Dataset], self._return_type)
+ if return_dataset_cls().get_model_class() is dataset.get_model_class():
+ return dataset
else:
- file_suffix = file_suffixes.pop()
- serializers = self._serializer_registry.\
- detect_tar_file_serializers_from_file_suffix(file_suffix)
- if len(serializers) == 0:
- self._log(f'No serializer for file suffix "{file_suffix}" can be'
- f'determined. Aborting restore.')
- else:
- self._log(f'Reading dataset from a gzipped tarpack at'
- f' "{os.path.abspath(tar_file_path)}"')
-
- serializer = serializers[0]
- with open(tar_file_path, 'rb') as tarfile_binary:
- dataset = serializer.deserialize(tarfile_binary.read())
- return_dataset_cls = cast(Type[Dataset], self._return_type)
- if return_dataset_cls().get_model_class(
- ) is dataset.get_model_class():
- return dataset
+ try:
+ new_dataset = return_dataset_cls()
+ if new_dataset.get_model_class() is Model[str]:
+ new_dataset.from_data(dataset.to_json())
else:
- try:
- new_dataset = return_dataset_cls()
- if new_dataset.get_model_class() is Model[str]:
- new_dataset.from_data(dataset.to_json())
- else:
- new_dataset.from_json(dataset.to_data())
- return new_dataset
- except:
- return dataset
+ new_dataset.from_json(dataset.to_data())
+ return new_dataset
+ except Exception:
+ return dataset
raise RuntimeError('No persisted output')
diff --git a/src/omnipy/config/job.py b/src/omnipy/config/job.py
index 97f0ab60..cbe0ae3d 100644
--- a/src/omnipy/config/job.py
+++ b/src/omnipy/config/job.py
@@ -1,10 +1,11 @@
from dataclasses import dataclass, field
-from datetime import datetime
+# from datetime import datetime
from pathlib import Path
-from typing import Optional
from omnipy.api.enums import ConfigPersistOutputsOptions, ConfigRestoreOutputsOptions
+# from typing import Optional
+
def _get_persist_data_dir_path() -> str:
return str(Path.cwd().joinpath(Path('data')))
diff --git a/src/omnipy/data/serializer.py b/src/omnipy/data/serializer.py
index 2c17421d..52c1158d 100644
--- a/src/omnipy/data/serializer.py
+++ b/src/omnipy/data/serializer.py
@@ -124,7 +124,7 @@ def _to_data_from_data_if_direct(dataset, serializer: Serializer):
try:
new_dataset = func(dataset, serializer)
return new_dataset, serializer
- except (TypeError, ValueError, ValidationError, AssertionError) as e:
+ except (TypeError, ValueError, ValidationError, AssertionError):
pass
return None, None
diff --git a/src/omnipy/engine/job_runner.py b/src/omnipy/engine/job_runner.py
index 07f7141f..08ec8721 100644
--- a/src/omnipy/engine/job_runner.py
+++ b/src/omnipy/engine/job_runner.py
@@ -16,7 +16,9 @@ def _register_job_state(self, job: IsJob, state: RunState) -> None:
if self._registry:
self._registry.set_job_state(job, state)
- def _decorate_result_with_job_finalization_detector(self, job: IsJob, job_result: object):
+ def _decorate_result_with_job_finalization_detector( # noqa: C901
+ self, job: IsJob, job_result: object):
+ # TODO: Simplify _decorate_result_with_job_finalization_detector()
if isinstance(job_result, GeneratorType):
job_result = cast(GeneratorType, job_result)
@@ -148,7 +150,7 @@ def _dag_flow_runner_call_func(*args: object, **kwargs: object) -> Any:
job_callback_accept_decorator(_dag_flow_decorator)
@staticmethod
- def default_dag_flow_run_decorator(dag_flow: IsDagFlow) -> Any:
+ def default_dag_flow_run_decorator(dag_flow: IsDagFlow) -> Any: # noqa: C901
def _inner_run_dag_flow(*args: object, **kwargs: object):
results = {}
result = None
diff --git a/src/omnipy/modules/fairtracks/create_filter.py b/src/omnipy/modules/fairtracks/create_filter.py
index dcbdffc1..6f994536 100644
--- a/src/omnipy/modules/fairtracks/create_filter.py
+++ b/src/omnipy/modules/fairtracks/create_filter.py
@@ -7,7 +7,7 @@
import requests
#########################################################
-### user parameters
+# user parameters
#######################################################
download_all_projects = False
download_all_cases = False
@@ -17,7 +17,7 @@
# NB very few annotations per case, get all
#######################################################
-###endpoints definition
+# endpoints definition
########################################################
projects_endpt = 'https://api.gdc.cancer.gov/projects'
cases_endpt = 'https://api.gdc.cancer.gov/cases'
@@ -73,7 +73,7 @@
projects_list = (response.json()['data']['hits'])
##########################################################################
-#Step3: filtered query on 'cases' (filter on project_id) to get cases ID for each TCGA project
+# Step3: filtered query on 'cases' (filter on project_id) to get cases ID for each TCGA project
# the IDs for all the files are also available form this endpoint
########################################################################
@@ -102,7 +102,7 @@
proj['cases'] = cases
##########################################################################
-#step4: filtered query on 'annotations' (filter on case_id) to get cases ID for each TCGA project
+# step4: filtered query on 'annotations' (filter on case_id) to get cases ID for each TCGA project
# the IDs for all the files are also available form this endpoint
########################################################################
diff --git a/src/omnipy/modules/fairtracks/functions.py b/src/omnipy/modules/fairtracks/functions.py
index f6b02256..f9e12e0b 100644
--- a/src/omnipy/modules/fairtracks/functions.py
+++ b/src/omnipy/modules/fairtracks/functions.py
@@ -23,12 +23,9 @@ def encode_api(endpoint='experiments', id=None, limit=None, format='json', frame
# ['projects', 'cases', 'files', 'annotations'], starting_point='0', size='25'
def gdc_api(object_type='projects', starting_point=None, size=None):
- api_url = GDC_BASE_URL + object_type + '/' + '?' + \
- '&'.join(
- (['from=' + starting_point] if starting_point else [])
- + (['size=' + size] if size else [])
- + (['expand=' + 'project'] if object_type == 'cases' else [])
- )
+ api_url = GDC_BASE_URL + object_type + '/' + '?' + '&'.join(
+ (['from=' + starting_point] if starting_point else []) + (['size=' + size] if size else [])
+ + ([+ 'project'] if object_type == 'cases' else []))
print(api_url)
response = requests.get(api_url)
if response.status_code != 200:
diff --git a/src/omnipy/modules/fairtracks/get_from_filter.py b/src/omnipy/modules/fairtracks/get_from_filter.py
index 3abe7e5e..ba9717b0 100644
--- a/src/omnipy/modules/fairtracks/get_from_filter.py
+++ b/src/omnipy/modules/fairtracks/get_from_filter.py
@@ -13,7 +13,7 @@
with open(endpt_str + '_filter.json', 'r') as infile:
filters = json.load(infile)
- params = { #'fields': fields,
+ params = { # 'fields': fields,
'filters': json.dumps(filters),
}
response = requests.get(endpt, params=params)
diff --git a/src/omnipy/modules/general/models.py b/src/omnipy/modules/general/models.py
index 7239cb8f..fadb3c8d 100644
--- a/src/omnipy/modules/general/models.py
+++ b/src/omnipy/modules/general/models.py
@@ -5,4 +5,4 @@
class NotIteratorExceptStrings(Model[object]):
def _parse_data(cls, data: object) -> object:
- assert isinstance(data, str) or not isinstance(data, Iterable)
\ No newline at end of file
+ assert isinstance(data, str) or not isinstance(data, Iterable)
diff --git a/src/omnipy/modules/general/tasks.py b/src/omnipy/modules/general/tasks.py
index c84e4a56..b062c2be 100644
--- a/src/omnipy/modules/general/tasks.py
+++ b/src/omnipy/modules/general/tasks.py
@@ -26,9 +26,13 @@ def split_dataset(
model_cls = dataset.get_model_class()
datafile_names_for_a = set(dataset.keys()) - set(datafile_names_for_b)
dataset_a = Dataset[model_cls]( # type: ignore
- {name: dataset[name] for name in dataset.keys() if name in datafile_names_for_a})
+ {
+ name: dataset[name] for name in dataset.keys() if name in datafile_names_for_a
+ })
dataset_b = Dataset[model_cls]( # type: ignore
- {name: dataset[name] for name in dataset.keys() if name in datafile_names_for_b})
+ {
+ name: dataset[name] for name in dataset.keys() if name in datafile_names_for_b
+ })
return dataset_a, dataset_b
diff --git a/src/omnipy/modules/pandas/__init__.py b/src/omnipy/modules/pandas/__init__.py
index ffd919bc..705596b3 100644
--- a/src/omnipy/modules/pandas/__init__.py
+++ b/src/omnipy/modules/pandas/__init__.py
@@ -1 +1 @@
-import pandas as pd
+import pandas as pd # noqa
diff --git a/src/omnipy/modules/prefect/__init__.py b/src/omnipy/modules/prefect/__init__.py
index 3ead6efd..805dc05b 100644
--- a/src/omnipy/modules/prefect/__init__.py
+++ b/src/omnipy/modules/prefect/__init__.py
@@ -20,6 +20,6 @@ def use_local_api_for_tests():
set_prefect_config_path()
use_local_api_for_tests()
-from prefect import flow, Flow, State, task, Task
-from prefect.tasks import task_input_hash
-from prefect.utilities.names import generate_slug
+from prefect import flow, Flow, State, task, Task # noqa
+from prefect.tasks import task_input_hash # noqa
+from prefect.utilities.names import generate_slug # noqa
diff --git a/src/omnipy/modules/raw/serializers.py b/src/omnipy/modules/raw/serializers.py
index ae7cc4f6..4219c4fc 100644
--- a/src/omnipy/modules/raw/serializers.py
+++ b/src/omnipy/modules/raw/serializers.py
@@ -1,17 +1,17 @@
from typing import Any, Dict, IO, Type
-from typing_inspect import get_generic_bases, get_generic_type, get_origin, get_parameters
-
from omnipy.data.dataset import Dataset
from omnipy.data.model import Model
from omnipy.data.serializer import TarFileSerializer
+# from typing_inspect import get_generic_bases, get_generic_type, get_origin, get_parameters
+
class RawDatasetToTarFileSerializer(TarFileSerializer):
""""""
@classmethod
def is_dataset_directly_supported(cls, dataset: Dataset) -> bool:
- return type(dataset) == Dataset[Model[str]]
+ return type(dataset) is Dataset[Model[str]]
@classmethod
def get_dataset_cls_for_new(cls) -> Type[Dataset]:
diff --git a/src/omnipy/util/callable_decorator_cls.py b/src/omnipy/util/callable_decorator_cls.py
index 26e982ae..d9bb33ff 100644
--- a/src/omnipy/util/callable_decorator_cls.py
+++ b/src/omnipy/util/callable_decorator_cls.py
@@ -6,7 +6,8 @@
from omnipy.api.types import DecoratorClassT
-def callable_decorator_cls(cls: Type[DecoratorClassT]) -> IsCallableClass[DecoratorClassT]:
+def callable_decorator_cls( # noqa: C901
+ cls: Type[DecoratorClassT]) -> IsCallableClass[DecoratorClassT]:
"""
"Meta-decorator" that allows any class to function as a decorator for a callable.
@@ -81,8 +82,8 @@ def _init(callable_arg: Callable) -> None:
_init(_callable_arg)
else:
# Add an instance-level _obj_call method, which are again callable by the
- # class-level __call__ method. When this method is called, the provided _callable_arg
- # is decorated.
+ # class-level __call__ method. When this method is called, the provided
+ # _callable_arg is decorated.
def _init_as_obj_call_method(
self, _callable_arg: Callable) -> Type[DecoratorClassT]: # noqa
_init(_callable_arg)
diff --git a/src/omnipy/util/mixin.py b/src/omnipy/util/mixin.py
index 3365c6dc..75d52c9e 100644
--- a/src/omnipy/util/mixin.py
+++ b/src/omnipy/util/mixin.py
@@ -144,7 +144,7 @@ def __new__(cls, *args, **kwargs):
return obj
@classmethod
- def _create_subcls_inheriting_from_mixins_and_orig_cls(cls):
+ def _create_subcls_inheriting_from_mixins_and_orig_cls(cls): # noqa: C901
# TODO: Refactor this, and possibly elsewhere in class
diff --git a/tests/compute/helpers/mocks.py b/tests/compute/helpers/mocks.py
index fd4bdfa0..504dfe03 100644
--- a/tests/compute/helpers/mocks.py
+++ b/tests/compute/helpers/mocks.py
@@ -37,13 +37,6 @@ def _call_job(self, *args: object, **kwargs: object) -> object:
...
-#
-# def mock_flow_template_callable_decorator_cls(
-# cls: Type['MockFlowTemplateSubclass']
-# ) -> IsFuncJobTemplateCallable['MockFlowTemplateSubclass']:
-# return cast(IsFuncJobTemplateCallable['MockFlowTemplateSubclass'], callable_decorator_cls(cls))
-
-
# @callable_decorator_cls
class MockFlowTemplateSubclass(JobTemplateMixin, JobBase):
@classmethod
@@ -289,7 +282,8 @@ def reset_persisted_time_of_cur_toplevel_flow_run(cls) -> None:
def _call_func(self, *args: object, **kwargs: object) -> object:
if self.persisted_time_of_cur_toplevel_flow_run:
- assert self.persisted_time_of_cur_toplevel_flow_run == self.time_of_cur_toplevel_flow_run
+ assert self.persisted_time_of_cur_toplevel_flow_run == \
+ self.time_of_cur_toplevel_flow_run
else:
self._persisted_time_of_cur_toplevel_flow_run.append(self.time_of_cur_toplevel_flow_run)
diff --git a/tests/compute/test_decorators.py b/tests/compute/test_decorators.py
index 3d053c91..67e5a642 100644
--- a/tests/compute/test_decorators.py
+++ b/tests/compute/test_decorators.py
@@ -43,7 +43,7 @@ def test_linear_flow_template_as_decorator(
plus_five_template: LinearFlowTemplate,
) -> None:
- assert (plus_five_template, LinearFlowTemplate)
+ assert isinstance(plus_five_template, LinearFlowTemplate)
assert plus_five_template.name == 'plus_five'
plus_five = plus_five_template.apply()
@@ -62,7 +62,7 @@ def test_dag_flow_template_as_decorator(
plus_five_template: DagFlowTemplate,
) -> None:
- assert (plus_five_template, DagFlowTemplate)
+ assert isinstance(plus_five_template, DagFlowTemplate)
assert plus_five_template.name == 'plus_five'
plus_five = plus_five_template.apply()
@@ -86,7 +86,7 @@ def test_func_flow_template_as_decorator(
plus_y_template: FuncFlowTemplate,
) -> None:
- assert (plus_y_template, FuncFlowTemplate)
+ assert isinstance(plus_y_template, FuncFlowTemplate)
assert plus_y_template.name == 'plus_y'
plus_y = plus_y_template.apply()
diff --git a/tests/engine/test_all_engines.py b/tests/engine/test_all_engines.py
index 39ab8009..9c2b8175 100644
--- a/tests/engine/test_all_engines.py
+++ b/tests/engine/test_all_engines.py
@@ -1,3 +1,5 @@
+import os
+
import pytest
import pytest_cases as pc
@@ -5,6 +7,11 @@
from .helpers.functions import run_job_test
+@pytest.mark.skipif(
+ os.getenv('OMNIPY_FORCE_SKIPPED_TEST') != '1',
+ reason="""
+TODO: Stopped working in some Prefect version between 2.10.10 and 2.13.3
+""")
@pc.parametrize(
'job_case',
[pc.fixture_ref('all_func_types_mock_jobs_all_engines_assert_runstate_mock_reg')],
diff --git a/tests/hub/test_runtime.py b/tests/hub/test_runtime.py
index d5feeed2..18205dde 100644
--- a/tests/hub/test_runtime.py
+++ b/tests/hub/test_runtime.py
@@ -1,4 +1,3 @@
-import logging
import os
from pathlib import Path
from typing import Annotated, Type
diff --git a/tests/integration/novel/full/test_multi_model_dataset.py b/tests/integration/novel/full/test_multi_model_dataset.py
index 6089bc41..fed9de06 100644
--- a/tests/integration/novel/full/test_multi_model_dataset.py
+++ b/tests/integration/novel/full/test_multi_model_dataset.py
@@ -16,11 +16,15 @@
def test_table_models():
- _a = GeneralTable([{'a': 123, 'b': 'ads'}, {'a': 234, 'b': 'acs'}])
- _b = TableTemplate[MyRecordSchema]([{'a': 123, 'b': 'ads'}, {'a': 234, 'b': 'acs'}])
+ _a = GeneralTable([{'a': 123, 'b': 'ads'}, {'a': 234, 'b': 'acs'}]) # noqa: F841
+ # yapf: disable
+ _b = TableTemplate[MyRecordSchema]([{'a': 123, 'b': 'ads'}, # noqa: F841
+ {'a': 234, 'b': 'acs'}])
with pytest.raises(ValidationError):
- _c = TableTemplate[MyOtherRecordSchema]([{'a': 123, 'b': 'ads'}, {'a': 234, 'b': 'acs'}])
+ _c = TableTemplate[MyOtherRecordSchema]([{'a': 123, 'b': 'ads'}, # noqa: F841
+ {'a': 234, 'b': 'acs'}])
+ # yapf: disable
# print(_a.to_json_schema(pretty=True))
# print(_b.to_json_schema(pretty=True))
@@ -138,4 +142,4 @@ def test_fail_run_specialize_record_models_inconsistent_types(
old_dataset['b'] = [{'b': 'df', 'c': True}, {'b': False, 'c': 'sg'}]
with pytest.raises(AssertionError):
- _new_dataset = specialize_record_models(old_dataset)
+ _new_dataset = specialize_record_models(old_dataset) # noqa: F841
diff --git a/tests/integration/novel/serialize/test_serialize.py b/tests/integration/novel/serialize/test_serialize.py
index 14f18d26..51db6716 100644
--- a/tests/integration/novel/serialize/test_serialize.py
+++ b/tests/integration/novel/serialize/test_serialize.py
@@ -8,7 +8,7 @@
PersistOutputsOptions,
RestoreOutputsOptions)
from omnipy.api.protocols.public.hub import IsRuntime
-from omnipy.compute.task import FuncArgJobBase, TaskTemplate
+from omnipy.compute.task import FuncArgJobBase
@pc.parametrize_with_cases('case_tmpl', cases='.cases.jobs', has_tag='task', prefix='case_config_')
diff --git a/tests/integration/reused/engine/conftest.py b/tests/integration/reused/engine/conftest.py
index ad730224..4251d05e 100644
--- a/tests/integration/reused/engine/conftest.py
+++ b/tests/integration/reused/engine/conftest.py
@@ -33,8 +33,11 @@ def all_job_classes(
dag_flow_template_cls: Type[IsDagFlowTemplate],
func_flow_template_cls: Type[IsFuncFlowTemplate],
):
- return job_type, task_template_cls, linear_flow_template_cls, \
- dag_flow_template_cls, func_flow_template_cls
+ return (job_type,
+ task_template_cls,
+ linear_flow_template_cls,
+ dag_flow_template_cls,
+ func_flow_template_cls)
@pc.fixture(scope='function', name='plain_engine')
@@ -65,7 +68,11 @@ def all_func_types_real_jobs_all_engines_real_reg(
engine_decorator: Optional[Callable[[IsEngine], IsEngine]],
registry: Optional[IsRunStateRegistry],
):
- job_type, task_template_cls, linear_flow_template_cls, dag_flow_template_cls, func_flow_template_cls = job_classes
+ (job_type,
+ task_template_cls,
+ linear_flow_template_cls,
+ dag_flow_template_cls,
+ func_flow_template_cls) = job_classes
# TODO: Fix job_type comparisons everywhere (bug due to pytest.fixture?)
if job_type.value == JobType.linear_flow.value:
diff --git a/tests/log/helpers/functions.py b/tests/log/helpers/functions.py
index c3614faa..fb1534d7 100644
--- a/tests/log/helpers/functions.py
+++ b/tests/log/helpers/functions.py
@@ -1,10 +1,7 @@
-from datetime import datetime
from io import StringIO
import os
from typing import List, Optional
-from omnipy.util.helpers import get_datetime_format
-
def read_log_lines_from_stream(str_stream: StringIO) -> List[str]:
str_stream.seek(0)
diff --git a/tests/modules/pandas/test_pandas.py b/tests/modules/pandas/test_pandas.py
index 9509bdbc..475b2cb9 100644
--- a/tests/modules/pandas/test_pandas.py
+++ b/tests/modules/pandas/test_pandas.py
@@ -72,8 +72,8 @@ def test_pandas_dataset_list_of_objects_different_keys():
@pytest.mark.skipif(
os.getenv('OMNIPY_FORCE_SKIPPED_TEST') != '1',
reason="""
-Pandas converts 'a' column into 'Int64' since all values can be cast into integers. Should remain
-floats.
+Pandas converts 'a' column into 'Int64' since all values can be cast into integers.
+ Should remain floats.
""")
def test_pandas_dataset_list_of_objects_float_numbers():
pandas_data = PandasDataset()
diff --git a/tests/modules/prefect/test_prefect.py b/tests/modules/prefect/test_prefect.py
index 8103632e..969391f8 100644
--- a/tests/modules/prefect/test_prefect.py
+++ b/tests/modules/prefect/test_prefect.py
@@ -3,7 +3,7 @@
def test_no_prefect_console_handler_in_root_logger():
- import omnipy.modules.prefect
+ import omnipy.modules.prefect # noqa: F401
assert os.environ['PREFECT_LOGGING_SETTINGS_PATH'].endswith('logging.yml')
@@ -14,7 +14,7 @@ def test_no_prefect_console_handler_in_root_logger():
def test_only_local_orion():
- import omnipy.modules.prefect
+ import omnipy.modules.prefect # noqa: F401
assert os.environ['PREFECT_API_KEY'] == ''
assert os.environ['PREFECT_API_URL'] == ''
diff --git a/tests/test_omnipy.py b/tests/test_omnipy.py
index 8a608e18..f7eefcff 100644
--- a/tests/test_omnipy.py
+++ b/tests/test_omnipy.py
@@ -2,4 +2,4 @@
def test_version():
- assert __version__ == '0.10.4'
+ assert __version__ == '0.10.5'
diff --git a/tests/util/test_callable_decorator_cls.py b/tests/util/test_callable_decorator_cls.py
index b8aa886f..6b14c594 100644
--- a/tests/util/test_callable_decorator_cls.py
+++ b/tests/util/test_callable_decorator_cls.py
@@ -58,7 +58,7 @@ def test_plain_decorator() -> None:
def my_func(*args: object, **kwargs: object) -> Dict[str, object]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -72,7 +72,7 @@ def test_plain_decorator_parentheses() -> None:
def my_func(*args: object, **kwargs: object) -> Dict[str, object]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -86,7 +86,7 @@ def test_decorator_with_kwargs() -> None:
def my_func(*args: object, **kwargs: object) -> Dict[str, object]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -100,7 +100,7 @@ def test_decorator_with_args_and_kwargs() -> None:
def my_func(*args: object, **kwargs: object) -> Dict[str, object]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -117,7 +117,7 @@ def other_func():
def my_func(*args: object, **kwargs: object) -> Dict[str, Any]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -132,7 +132,7 @@ def test_double_decorator_with_args_and_kwargs() -> None:
def my_func(*args: object, **kwargs: object) -> Dict[str, Any]:
return dict(args=args, kwargs=kwargs)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -167,7 +167,7 @@ def my_fancy_func(*args: Union[int, str], **kwargs: bool) -> Dict[str, Any]:
def test_decorator_as_function_fancy_func() -> None:
my_func = MockClass(my_fancy_func)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -179,7 +179,7 @@ def test_decorator_as_function_fancy_func() -> None:
def test_decorator_as_function_fancy_func_args_kwargs() -> None:
my_func = MockClass(123, True, param=123, other=True)(my_fancy_func)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
_assert_func_decoration(my_func) # noqa
_assert_call_func(my_func)
@@ -191,7 +191,7 @@ def test_decorator_as_function_fancy_func_args_kwargs() -> None:
def test_fail_decorator_as_function_no_args_no_func() -> None:
my_func = MockClass()
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
with pytest.raises(TypeError):
my_func() # type: ignore # noqa
@@ -204,7 +204,7 @@ def test_fail_decorator_as_function_no_args_no_func() -> None:
def test_fail_decorator_as_function_args_and_kwargs_no_func() -> None:
my_func = MockClass(123, True, param=123, other=True)
- assert type(my_func) == MockClass
+ assert type(my_func) is MockClass
with pytest.raises(TypeError):
my_func()
diff --git a/tests/util/test_dynamic_mixin.py b/tests/util/test_dynamic_mixin.py
index bd9b58c8..d51cdd2c 100644
--- a/tests/util/test_dynamic_mixin.py
+++ b/tests/util/test_dynamic_mixin.py
@@ -1,4 +1,4 @@
-from abc import ABCMeta
+# from abc import ABCMeta
from inspect import Parameter, signature
from typing import Dict, Generic, get_args, Optional, Tuple, Type, TypeVar
@@ -114,7 +114,7 @@ def override(self):
class MockKwArgAccessOrigClsMemberStateMixin:
def __init__(self, *, my_kwarg):
- self._value = f'my_kwarg: {my_kwarg}' if self.kwargs.get('verbose') == True else my_kwarg
+ self._value = f'my_kwarg: {my_kwarg}' if self.kwargs.get('verbose', False) else my_kwarg
def new_method(self):
return self._value
@@ -182,7 +182,7 @@ def _assert_args_and_kwargs(mock_obj: object,
if mock_cls:
assert tuple(signature(mock_cls.__init__).parameters.keys()) == \
- tuple(['self'] + non_self_param_keys)
+ tuple(['self'] + non_self_param_keys)
def test_new_method_no_state_mixin(mock_plain_cls):
@@ -513,7 +513,7 @@ def override(self):
def test_predefined_init_kwargs_progressive_multiple_state_mixins_same_default(
mock_predefined_init_kwargs_cls):
- MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls #noqa
+ MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls # noqa
MockPredefinedInitKwargsCls.accept_mixin(MockKwArgStateMixin)
@@ -561,7 +561,7 @@ def test_predefined_init_kwargs_progressive_multiple_state_mixins_same_default(
def test_predefined_init_kwargs_progressive_multiple_state_mixins_different_withmixin_classes(
mock_predefined_init_kwargs_cls):
- MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls #noqa
+ MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls # noqa
MockPredefinedInitKwargsCls.accept_mixin(MockKwArgStateMixin)
@@ -602,7 +602,7 @@ def test_predefined_init_kwargs_progressive_multiple_state_mixins_different_with
def test_nested_mixins(mock_predefined_init_kwargs_cls, mock_plain_cls):
- MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls #noqa
+ MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls # noqa
MockPlainCls = mock_plain_cls # noqa
MockPredefinedInitKwargsCls.accept_mixin(MockKwArgStateMixin)
@@ -625,7 +625,7 @@ def test_nested_mixins(mock_predefined_init_kwargs_cls, mock_plain_cls):
def test_nested_mixins_static_outer_inheritance(mock_predefined_init_kwargs_cls):
- MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls #noqa
+ MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls # noqa
MockPredefinedInitKwargsCls.accept_mixin(MockKwArgStateMixin)
class MockMockPlainCls(MockPredefinedInitKwargsCls):
@@ -654,7 +654,7 @@ def override(self):
def test_nested_mixins_double_static_outer_inheritance(mock_predefined_init_kwargs_cls):
- MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls #noqa
+ MockPredefinedInitKwargsCls = mock_predefined_init_kwargs_cls # noqa
MockPredefinedInitKwargsCls.accept_mixin(MockKwArgStateMixin)
class MockMockPlainCls(MockPredefinedInitKwargsCls):
@@ -687,7 +687,7 @@ class MockMockMockPlainCls(MockMockPlainCls):
def test_nested_mixins_static_outer_inheritance_from_generic(
mock_predefined_init_kwargs_generic_cls):
- MockPredefinedInitKwargsGenericCls = mock_predefined_init_kwargs_generic_cls #noqa
+ MockPredefinedInitKwargsGenericCls = mock_predefined_init_kwargs_generic_cls # noqa
MockPredefinedInitKwargsGenericCls.accept_mixin(MockKwArgStateMixin)
class MockMockPlainCls(MockPredefinedInitKwargsGenericCls[int]):
diff --git a/tests/util/test_helpers.py b/tests/util/test_helpers.py
index 2f292821..9911473c 100644
--- a/tests/util/test_helpers.py
+++ b/tests/util/test_helpers.py
@@ -1,6 +1,6 @@
-from typing import Dict, Generic, get_args, get_type_hints, TypeVar
+from typing import Dict, Generic, get_args, TypeVar
-from typing_inspect import get_args, get_bound, get_generic_type, get_last_args, get_parameters
+from typing_inspect import get_generic_type
from omnipy.util.helpers import transfer_generic_args_to_cls