-
Notifications
You must be signed in to change notification settings - Fork 22
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
nsviewer fails to display an image properly #15
Comments
|
But it looks so pretty! You sure you want to fix it? Is the image 91 x 109 x 91? If not, that's probably the issue. Currently, transformations between image/world/canvas spaces are hardcoded to only work for those dimensions. This needs to be fixed by generalizing the matrix transformations in the Transform class in models.coffee to handle arbitrary dimensions based on the header. |
|
Is this happening when loading from json, from nifti, or both? |
|
JSON. Does the viewer support loading niftis directly? We can get rid of On Mon, Oct 28, 2013 at 6:05 PM, Tal Yarkoni [email protected]:
|
|
Yes, the viewer supports loading nifti directly--this was one of the things we did at the hackathon in Boulder, remember? ;) The tradeoff between the json and nifti approaches amounts to server vs. client-side loading time right now. For reasons I'm not entirely clear on, the xtk I/O code (which is what I'm using to read Nifti images in) is quite slow compared to constructing a volume from json. This is presumably an implementation issue, since reading from binary with typed arrays should be faster than filling a volume from plain text. But right now most json images load in under a second, whereas larger nifti volumes can take a few seconds to load from binary. But given that reading directly from nifti pushes the load to the client, and eliminates the need for server-side conversion, I would also support moving to binary-only. That's actually how neurosynth.org has been operating for the past few months. The nsviewer documentation/example is already loading the MNI template from nifti, so you can look at that to see how it's done. Actually, I'm pretty sure that if you just change the filename from .json to .nii.gz or .nii everything should work seamlessly without having to do anything else. |
This image https://dl.dropboxusercontent.com/u/412959/spmT_0001.nii is displayed as a scrambled matrix.
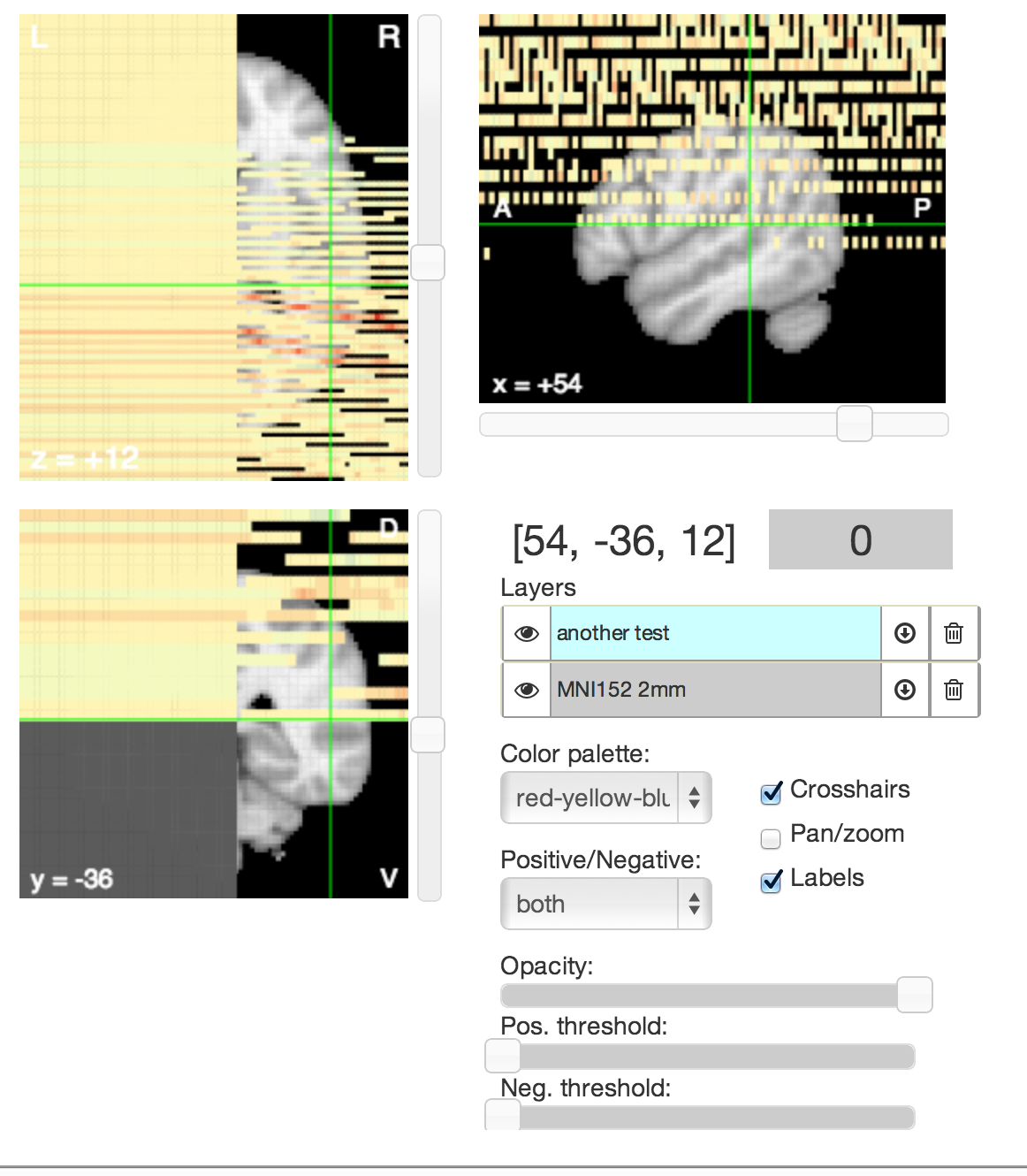
The text was updated successfully, but these errors were encountered: