diff --git a/.dockerignore b/.dockerignore
new file mode 100644
index 0000000..7448793
--- /dev/null
+++ b/.dockerignore
@@ -0,0 +1,3 @@
+work_dirs/
+artifacts/
+wandb/
diff --git a/.github/ISSUE_TEMPLATE/bug_report.md b/.github/ISSUE_TEMPLATE/bug_report.md
new file mode 100644
index 0000000..c00c1f5
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/bug_report.md
@@ -0,0 +1,42 @@
+---
+name: Bug report
+about: Create a report to help us improve
+title: '[Bug]'
+labels: bug
+assignees: ''
+---
+
+### Describe the bug
+
+A clear and concise description of what the bug is.
+
+\[here\]
+
+### To Reproduce
+
+The command you executed.
+
+```shell
+[here]
+```
+
+### Post related information
+
+1. The output of `pip list | grep "mmcv\|mmcls\|^torch"`
+ \[here\]
+2. Your config file if you modified it or created a new one.
+
+```python
+[here]
+```
+
+3. Your train log file if you meet the problem during training.
+ \[here\]
+4. Other code you modified in the `mmcls` folder.
+ \[here\]
+
+### Additional context
+
+Add any other context about the problem here.
+
+\[here\]
diff --git a/.github/ISSUE_TEMPLATE/feature_request.md b/.github/ISSUE_TEMPLATE/feature_request.md
new file mode 100644
index 0000000..23b7c09
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/feature_request.md
@@ -0,0 +1,32 @@
+---
+name: Feature request
+about: Suggest an idea for this project
+title: '[Feature]'
+labels: enhancement
+assignees: ''
+---
+
+### Describe the feature
+
+\[here\]
+
+### Motivation
+
+A clear and concise description of the motivation of the feature.
+Ex1. It is inconvenient when \[....\].
+Ex2. There is a recent paper \[....\], which is very helpful for \[....\].
+
+\[here\]
+
+### Related resources
+
+If there is an official code release or third-party implementation, please also provide the information here, which would be very helpful.
+
+\[here\]
+
+### Additional context
+
+Add any other context or screenshots about the feature request here.
+If you would like to implement the feature and create a PR, please leave a comment here and that would be much appreciated.
+
+\[here\]
diff --git a/.github/ISSUE_TEMPLATE/general-questions.md b/.github/ISSUE_TEMPLATE/general-questions.md
new file mode 100644
index 0000000..42d5fb2
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/general-questions.md
@@ -0,0 +1,31 @@
+---
+name: General questions
+about: 'Ask general questions to get help '
+title: ''
+labels: help wanted
+assignees: ''
+---
+
+### Checklist
+
+- I have searched related issues but cannot get the expected help.
+- I have read related documents and don't know what to do.
+
+### Describe the question you meet
+
+\[here\]
+
+### Post related information
+
+1. The output of `pip list | grep "mmcv\|mmcls\|^torch"`
+ \[here\]
+2. Your config file if you modified it or created a new one.
+
+```python
+[here]
+```
+
+3. Your train log file if you meet the problem during training.
+ \[here\]
+4. Other code you modified in the `mmcls` folder.
+ \[here\]
diff --git a/.github/pull_request_template.md b/.github/pull_request_template.md
new file mode 100644
index 0000000..3916457
--- /dev/null
+++ b/.github/pull_request_template.md
@@ -0,0 +1,28 @@
+Thanks for your contribution and we appreciate it a lot. The following instructions would make your pull request more healthy and more easily get feedback. If you do not understand some items, don't worry, just make the pull request and seek help from maintainers.
+
+## Motivation
+
+Please describe the motivation of this PR and the goal you want to achieve through this PR.
+
+## Modification
+
+Please briefly describe what modification is made in this PR.
+
+## Results (Optional)
+
+| Dataset | Model | box AP |
+| :-----: | :---: | :----: |
+| | | |
+
+## Use cases (Optional)
+
+If this PR introduces a new feature, it is better to list some use cases here and update the documentation.
+
+## Checklist
+
+**Before PR**:
+
+- [ ] Pre-commit or other linting tools are used to fix the potential lint issues.
+- [ ] Bug fixes are fully covered by unit tests, the case that causes the bug should be added in the unit tests.
+- [ ] The modification is covered by complete unit tests. If not, please add more unit test to ensure the correctness.
+- [ ] The documentation has been modified accordingly, like docstring or example tutorials.
diff --git a/.github/workflows/build.yml b/.github/workflows/build.yml
new file mode 100644
index 0000000..ea40c59
--- /dev/null
+++ b/.github/workflows/build.yml
@@ -0,0 +1,66 @@
+name: build
+
+on: [pull_request]
+
+jobs:
+ lint:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v2
+ - name: Set up Python 3.7
+ uses: actions/setup-python@v2
+ with:
+ python-version: 3.8
+ - name: Install pre-commit hook
+ run: |
+ pip install pre-commit
+ pre-commit install
+ - name: Linting
+ run: pre-commit run --all-files
+ - name: Check docstring coverage
+ run: |
+ pip install interrogate
+ interrogate -v --ignore-init-method --ignore-module --ignore-nested-functions --ignore-regex "__repr__" --fail-under 0 dethub
+
+ build_cuda:
+ runs-on: ubuntu-18.04
+ env:
+ NVIDIA_DRIVER_CAPABILITIES: 'utility,compute'
+ DEBIAN_FRONTEND: noninteractive
+ container:
+ image: nvcr.io/nvidia/pytorch:22.07-py3
+ strategy:
+ matrix:
+ include:
+ - mmcv: 1.6.0
+ mmcv_link: "cu115/torch1.11.0"
+
+ steps:
+ - uses: actions/checkout@v2
+ - name: Install utils
+ run: pip install psutil
+ - name: Install system dependencies
+ run: |
+ apt-get update && apt-get install -y vim libgl1-mesa-dev
+ apt-get clean
+ rm -rf /var/lib/apt/lists/*
+ FORCE_CUDA="1"
+ - name: Install mmdet dependencies
+ run: |
+ pip install setuptools
+ pip install --no-cache-dir openmim==0.3.0 && \
+ pip install -r requirements/requirements.txt
+ pip install -r requirements/tests.txt
+ pip uninstall -y opencv-python && pip install opencv-python==4.5.1.48 && \
+ MMCV_WITH_OPS=1 pip install mmcv==2.0.0rc1 && \
+ pip install 'git+https://github.com/facebookresearch/detectron2.git'
+ python -c "import mmcv"
+ - name: Build and install
+ run: rm -rf .eggs && pip install -e .
+ - name: Soft link
+ run: ln -s /opt/conda/lib/python3.8/site-packages /opt/site-packages
+ - name: Run unittests and generate coverage report
+ run: |
+ coverage run --branch --source=dethub -m pytest tests/
+ coverage xml
+ coverage report -m
diff --git a/.gitignore b/.gitignore
new file mode 100644
index 0000000..4fa2e23
--- /dev/null
+++ b/.gitignore
@@ -0,0 +1,173 @@
+### https://raw.github.com/github/gitignore/218a941be92679ce67d0484547e3e142b2f5f6f0/Global/macOS.gitignore
+
+# General
+.DS_Store
+.AppleDouble
+.LSOverride
+
+# Icon must end with two \r
+Icon
+
+
+# Thumbnails
+._*
+
+# Files that might appear in the root of a volume
+.DocumentRevisions-V100
+.fseventsd
+.Spotlight-V100
+.TemporaryItems
+.Trashes
+.VolumeIcon.icns
+.com.apple.timemachine.donotpresent
+
+# Directories potentially created on remote AFP share
+.AppleDB
+.AppleDesktop
+Network Trash Folder
+Temporary Items
+.apdisk
+
+
+### https://raw.github.com/github/gitignore/218a941be92679ce67d0484547e3e142b2f5f6f0/Python.gitignore
+
+# Byte-compiled / optimized / DLL files
+__pycache__/
+*.py[cod]
+*$py.class
+
+# C extensions
+*.so
+
+# Distribution / packaging
+.Python
+build/
+develop-eggs/
+dist/
+downloads/
+eggs/
+.eggs/
+lib/
+lib64/
+parts/
+sdist/
+var/
+wheels/
+share/python-wheels/
+*.egg-info/
+.installed.cfg
+*.egg
+MANIFEST
+
+# PyInstaller
+# Usually these files are written by a python script from a template
+# before PyInstaller builds the exe, so as to inject date/other infos into it.
+*.manifest
+*.spec
+
+# Installer logs
+pip-log.txt
+pip-delete-this-directory.txt
+
+# Unit test / coverage reports
+htmlcov/
+.tox/
+.nox/
+.coverage
+.coverage.*
+.cache
+nosetests.xml
+coverage.xml
+*.cover
+*.py,cover
+.hypothesis/
+.pytest_cache/
+cover/
+
+# Translations
+*.mo
+*.pot
+
+# Django stuff:
+*.log
+local_settings.py
+db.sqlite3
+db.sqlite3-journal
+
+# Flask stuff:
+instance/
+.webassets-cache
+
+# Scrapy stuff:
+.scrapy
+
+# Sphinx documentation
+docs/_build/
+
+# PyBuilder
+.pybuilder/
+target/
+
+# Jupyter Notebook
+.ipynb_checkpoints
+
+# IPython
+profile_default/
+ipython_config.py
+
+# pyenv
+# For a library or package, you might want to ignore these files since the code is
+# intended to run in multiple environments; otherwise, check them in:
+# .python-version
+
+# pipenv
+# According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
+# However, in case of collaboration, if having platform-specific dependencies or dependencies
+# having no cross-platform support, pipenv may install dependencies that don't work, or not
+# install all needed dependencies.
+#Pipfile.lock
+
+# PEP 582; used by e.g. github.com/David-OConnor/pyflow
+__pypackages__/
+
+# Celery stuff
+celerybeat-schedule
+celerybeat.pid
+
+# SageMath parsed files
+*.sage.py
+
+# Environments
+.env
+.venv
+env/
+venv/
+ENV/
+env.bak/
+venv.bak/
+
+# Spyder project settings
+.spyderproject
+.spyproject
+
+# Rope project settings
+.ropeproject
+
+# mkdocs documentation
+/site
+
+# mypy
+.mypy_cache/
+.dmypy.json
+dmypy.json
+
+# Pyre type checker
+.pyre/
+
+# pytype static type analyzer
+.pytype/
+
+# Cython debug symbols
+cython_debug/
+
+.idea
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 0000000..13a2d96
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,45 @@
+repos:
+ - repo: https://github.com/PyCQA/flake8
+ rev: 4.0.1
+ hooks:
+ - id: flake8
+ - repo: https://github.com/PyCQA/isort
+ rev: 5.10.1
+ hooks:
+ - id: isort
+ - repo: https://github.com/pre-commit/mirrors-yapf
+ rev: v0.30.0
+ hooks:
+ - id: yapf
+ - repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v3.1.0
+ hooks:
+ - id: trailing-whitespace
+ - id: check-yaml
+ - id: end-of-file-fixer
+ - id: requirements-txt-fixer
+ - id: double-quote-string-fixer
+ - id: check-merge-conflict
+ - id: fix-encoding-pragma
+ args: ["--remove"]
+ - id: mixed-line-ending
+ args: ["--fix=lf"]
+ - repo: https://github.com/executablebooks/mdformat
+ rev: 0.7.9
+ hooks:
+ - id: mdformat
+ args: ["--number", "--table-width", "200"]
+ additional_dependencies:
+ - mdformat-openmmlab
+ - mdformat_frontmatter
+ - linkify-it-py
+ - repo: https://github.com/codespell-project/codespell
+ rev: v2.1.0
+ hooks:
+ - id: codespell
+ args: ["--skip", "custom_det/core/evaluation/ot_cost.py"]
+ - repo: https://github.com/myint/docformatter
+ rev: v1.3.1
+ hooks:
+ - id: docformatter
+ args: ["--in-place", "--wrap-descriptions", "79"]
diff --git a/Dockerfile b/Dockerfile
new file mode 100644
index 0000000..db8102a
--- /dev/null
+++ b/Dockerfile
@@ -0,0 +1,27 @@
+FROM nvcr.io/nvidia/pytorch:22.07-py3
+
+RUN apt update -y && apt install -y \
+ git
+RUN apt-get update && apt-get install -y \
+ vim \
+ libgl1-mesa-dev
+ENV FORCE_CUDA="1"
+
+# Install python package.
+WORKDIR /dethub
+COPY ./ /dethub
+RUN pip install --upgrade pip && \
+ pip install --no-cache-dir openmim==0.3.1 && \
+ pip install . && \
+ pip uninstall -y opencv-python && pip install opencv-python==4.5.1.48 && \
+ MMCV_WITH_OPS=1 pip install mmcv==2.0.0rc1 && \
+ pip install 'git+https://github.com/facebookresearch/detectron2.git'
+
+# Language settings
+ENV LANG C.UTF-8
+ENV LANGUAGE en_US
+
+RUN ln -s /opt/conda/lib/python3.8/site-packages /opt/site-packages
+RUN git config --global --add safe.directory /workspace
+
+WORKDIR /workspace
diff --git a/MANIFEST.in b/MANIFEST.in
new file mode 100644
index 0000000..390c37d
--- /dev/null
+++ b/MANIFEST.in
@@ -0,0 +1,4 @@
+include fovkrt/*.txt
+include dethub/VERSION
+recursive-include dethub/.mim/configs *.py *.yml
+recursive-include dethub/.mim/tools *.sh *.py
diff --git a/README.md b/README.md
index f68aff5..8ddf386 100644
--- a/README.md
+++ b/README.md
@@ -1 +1,64 @@
-# open-detection-experiments
\ No newline at end of file
+# DetHub
+
+[](https://github.com/okotaku/dethub/actions/workflows/build.yml)
+[](https://github.com/okotaku/dethub/blob/main/LICENSE)
+
+## Introduction
+
+DetHub is an open source object detection / instance segmentation experiments hub. Our main contribution is supporting detection datasets and share baselines.
+
+- Support more and more datasets
+- Provide reproducible baseline configs for these datasets
+- Provide pretrained models, results and inference codes for these datasets
+
+Documentation: [docs](docs)
+
+## Supported Datasets
+
+- [x] [TensorFlow - Help Protect the Great Barrier Reef (Kaggle)](configs/projects/gbr_cots/)
+- [x] [LIVECell dataset](configs/projects/livecell/)
+- [x] [Sartorius - Cell Instance Segmentation (Kaggle)](configs/projects/sartorius_cellseg/)
+
+## Get Started
+
+Please refer to [get_started.md](docs/source/get_started.md) for get started.
+Other tutorials for:
+
+- [run](docs/source/run.md)
+
+## Contributing
+
+### CONTRIBUTING
+
+We appreciate all contributions to improve dethub. Please refer to [CONTRIBUTING.md](https://github.com/open-mmlab/mmcv/blob/master/CONTRIBUTING.md) for the contributing guideline.
+
+## License
+
+This project is released under the [Apache 2.0 license](LICENSE).
+
+## Acknowledgement
+
+This repo borrows the architecture design and part of the code from [mmdetection](https://github.com/open-mmlab/mmdetection).
+
+Also, please check the following openmmlab projects and the corresponding Documentation.
+
+- [OpenMMLab](https://openmmlab.com/)
+- [MMCV](https://github.com/open-mmlab/mmcv): OpenMMLab foundational library for computer vision.
+- [MIM](https://github.com/open-mmlab/mim): MIM Installs OpenMMLab Packages.
+- [MMClassification](https://github.com/open-mmlab/mmclassification): OpenMMLab image classification toolbox and benchmark.
+
+#### Citation
+
+```
+@article{mmdetection,
+ title = {{MMDetection}: Open MMLab Detection Toolbox and Benchmark},
+ author = {Chen, Kai and Wang, Jiaqi and Pang, Jiangmiao and Cao, Yuhang and
+ Xiong, Yu and Li, Xiaoxiao and Sun, Shuyang and Feng, Wansen and
+ Liu, Ziwei and Xu, Jiarui and Zhang, Zheng and Cheng, Dazhi and
+ Zhu, Chenchen and Cheng, Tianheng and Zhao, Qijie and Li, Buyu and
+ Lu, Xin and Zhu, Rui and Wu, Yue and Dai, Jifeng and Wang, Jingdong
+ and Shi, Jianping and Ouyang, Wanli and Loy, Chen Change and Lin, Dahua},
+ journal= {arXiv preprint arXiv:1906.07155},
+ year={2019}
+}
+```
diff --git a/configs/_base_/models/yolox_s.py b/configs/_base_/models/yolox_s.py
new file mode 100644
index 0000000..a74e857
--- /dev/null
+++ b/configs/_base_/models/yolox_s.py
@@ -0,0 +1,62 @@
+model = dict(
+ type='YOLOX',
+ data_preprocessor=dict(
+ type='DetDataPreprocessor',
+ pad_size_divisor=32,
+ batch_augments=[
+ dict(
+ type='BatchSyncRandomResize',
+ random_size_range=(480, 800),
+ size_divisor=32,
+ interval=10)
+ ]),
+ backbone=dict(
+ type='CSPDarknet',
+ deepen_factor=0.33,
+ widen_factor=0.5,
+ out_indices=(2, 3, 4),
+ use_depthwise=False,
+ spp_kernal_sizes=(5, 9, 13),
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish'),
+ ),
+ neck=dict(
+ type='YOLOXPAFPN',
+ in_channels=[128, 256, 512],
+ out_channels=128,
+ num_csp_blocks=1,
+ use_depthwise=False,
+ upsample_cfg=dict(scale_factor=2, mode='nearest'),
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish')),
+ bbox_head=dict(
+ type='YOLOXHead',
+ num_classes=80,
+ in_channels=128,
+ feat_channels=128,
+ stacked_convs=2,
+ strides=(8, 16, 32),
+ use_depthwise=False,
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish'),
+ loss_cls=dict(
+ type='CrossEntropyLoss',
+ use_sigmoid=True,
+ reduction='sum',
+ loss_weight=1.0),
+ loss_bbox=dict(
+ type='IoULoss',
+ mode='square',
+ eps=1e-16,
+ reduction='sum',
+ loss_weight=5.0),
+ loss_obj=dict(
+ type='CrossEntropyLoss',
+ use_sigmoid=True,
+ reduction='sum',
+ loss_weight=1.0),
+ loss_l1=dict(type='L1Loss', reduction='sum', loss_weight=1.0)),
+ train_cfg=dict(assigner=dict(type='SimOTAAssigner', center_radius=2.5)),
+ # In order to align the source code, the threshold of the val phase is
+ # 0.01, and the threshold of the test phase is 0.001.
+ test_cfg=dict(score_thr=0.01, nms=dict(type='nms', iou_threshold=0.65)))
diff --git a/configs/_base_/models/yolox_x.py b/configs/_base_/models/yolox_x.py
new file mode 100644
index 0000000..ed5deda
--- /dev/null
+++ b/configs/_base_/models/yolox_x.py
@@ -0,0 +1,62 @@
+model = dict(
+ type='YOLOX',
+ data_preprocessor=dict(
+ type='DetDataPreprocessor',
+ pad_size_divisor=32,
+ batch_augments=[
+ dict(
+ type='BatchSyncRandomResize',
+ random_size_range=(480, 800),
+ size_divisor=32,
+ interval=10)
+ ]),
+ backbone=dict(
+ type='CSPDarknet',
+ deepen_factor=1.33,
+ widen_factor=1.25,
+ out_indices=(2, 3, 4),
+ use_depthwise=False,
+ spp_kernal_sizes=(5, 9, 13),
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish'),
+ ),
+ neck=dict(
+ type='YOLOXPAFPN',
+ in_channels=[320, 640, 1280],
+ out_channels=320,
+ num_csp_blocks=4,
+ use_depthwise=False,
+ upsample_cfg=dict(scale_factor=2, mode='nearest'),
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish')),
+ bbox_head=dict(
+ type='YOLOXHead',
+ num_classes=80,
+ in_channels=320,
+ feat_channels=320,
+ stacked_convs=2,
+ strides=(8, 16, 32),
+ use_depthwise=False,
+ norm_cfg=dict(type='BN', momentum=0.03, eps=0.001),
+ act_cfg=dict(type='Swish'),
+ loss_cls=dict(
+ type='CrossEntropyLoss',
+ use_sigmoid=True,
+ reduction='sum',
+ loss_weight=1.0),
+ loss_bbox=dict(
+ type='IoULoss',
+ mode='square',
+ eps=1e-16,
+ reduction='sum',
+ loss_weight=5.0),
+ loss_obj=dict(
+ type='CrossEntropyLoss',
+ use_sigmoid=True,
+ reduction='sum',
+ loss_weight=1.0),
+ loss_l1=dict(type='L1Loss', reduction='sum', loss_weight=1.0)),
+ train_cfg=dict(assigner=dict(type='SimOTAAssigner', center_radius=2.5)),
+ # In order to align the source code, the threshold of the val phase is
+ # 0.01, and the threshold of the test phase is 0.001.
+ test_cfg=dict(score_thr=0.01, nms=dict(type='nms', iou_threshold=0.65)))
diff --git a/configs/projects/gbr_cots/README.md b/configs/projects/gbr_cots/README.md
new file mode 100644
index 0000000..fb65997
--- /dev/null
+++ b/configs/projects/gbr_cots/README.md
@@ -0,0 +1,60 @@
+# TensorFlow - Help Protect the Great Barrier Reef (Kaggle)
+
+Kaggle [TensorFlow - Help Protect the Great Barrier Reef](https://www.kaggle.com/competitions/tensorflow-great-barrier-reef)
+
+## Run demo
+
+```
+$ docker compose exec dethub python tools/image_demo.py configs/projects/gbr_cots/demo/5756.jpg configs/projects/gbr_cots/yolox/yolox_s_gbr_cots.py https://github.com/okotaku/dethub-weights/releases/download/v0.0.1/yolox_s_gbr_cots-cefaa435.pth --out-file configs/projects/gbr_cots/demo/5756_demo.jpg
+```
+
+
+
+## Prepare datasets
+
+1. Download competition data from Kaggle
+
+```
+kaggle competitions download -c tensorflow-great-barrier-reef
+```
+
+2. Download coco format json.
+
+```
+kaggle datasets download https://www.kaggle.com/datasets/takuok/gbrcotscocoformat
+```
+
+\*We prepared coco format files from [this script](../../../tools/dataset_converters/prepare_gbr_cots.py).
+
+3. Unzip the files as follows
+
+```
+data/gbr_cots
+├── train_images
+├── train.csv
+├── dtrain_g0.json
+└── dval_g0.json
+```
+
+## Run train
+
+Set env variables
+
+```
+$ export DATA_DIR=/path/to/data
+```
+
+Start a docker container
+
+```
+$ docker compose up -d dethub
+```
+
+Run train
+
+```
+# single gpu
+$ docker compose exec dethub python /opt/site-packages/mmdet/.mim/tools/train.py configs/projects/gbr_cots/yolox/yolox_s_gbr_cots.py
+# multi gpus
+$ docker compose exec dethub python -m torch.distributed.launch --nproc_per_node=2 /opt/site-packages/mmdet/.mim/tools/train.py configs/projects/gbr_cots/yolox/yolox_s_gbr_cots.py --launcher pytorch
+```
diff --git a/configs/projects/gbr_cots/demo/5756.jpg b/configs/projects/gbr_cots/demo/5756.jpg
new file mode 100644
index 0000000..a66df4a
Binary files /dev/null and b/configs/projects/gbr_cots/demo/5756.jpg differ
diff --git a/configs/projects/gbr_cots/demo/5756_demo.jpg b/configs/projects/gbr_cots/demo/5756_demo.jpg
new file mode 100644
index 0000000..697100f
Binary files /dev/null and b/configs/projects/gbr_cots/demo/5756_demo.jpg differ
diff --git a/configs/projects/gbr_cots/yolox/README.md b/configs/projects/gbr_cots/yolox/README.md
new file mode 100644
index 0000000..2a59ceb
--- /dev/null
+++ b/configs/projects/gbr_cots/yolox/README.md
@@ -0,0 +1,31 @@
+# YOLOX
+
+> [YOLOX: Exceeding YOLO Series in 2021](https://arxiv.org/abs/2107.08430)
+
+
+
+## Abstract
+
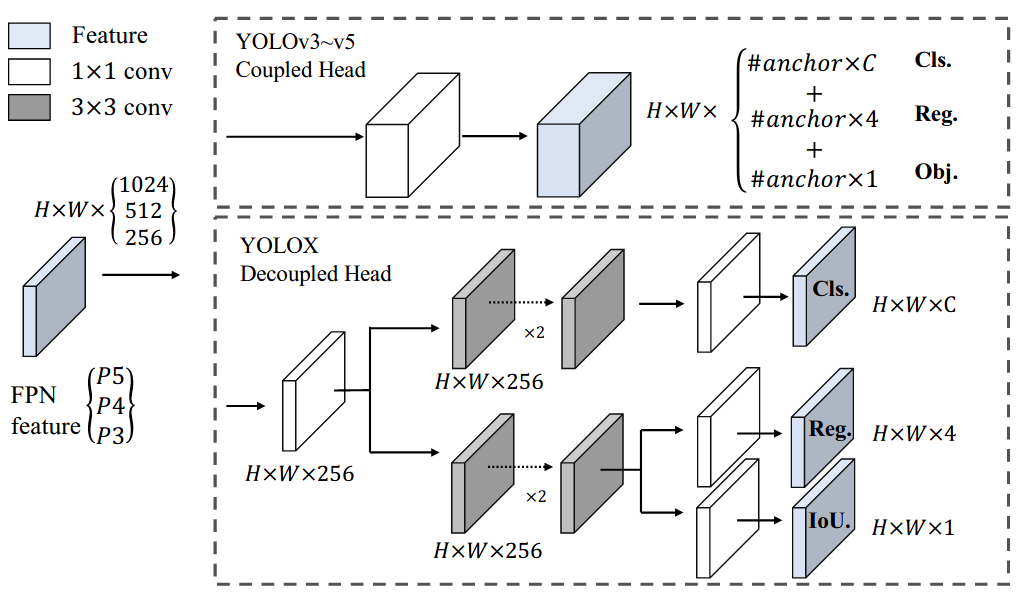
+In this report, we present some experienced improvements to YOLO series, forming a new high-performance detector -- YOLOX. We switch the YOLO detector to an anchor-free manner and conduct other advanced detection techniques, i.e., a decoupled head and the leading label assignment strategy SimOTA to achieve state-of-the-art results across a large scale range of models: For YOLO-Nano with only 0.91M parameters and 1.08G FLOPs, we get 25.3% AP on COCO, surpassing NanoDet by 1.8% AP; for YOLOv3, one of the most widely used detectors in industry, we boost it to 47.3% AP on COCO, outperforming the current best practice by 3.0% AP; for YOLOX-L with roughly the same amount of parameters as YOLOv4-CSP, YOLOv5-L, we achieve 50.0% AP on COCO at a speed of 68.9 FPS on Tesla V100, exceeding YOLOv5-L by 1.8% AP. Further, we won the 1st Place on Streaming Perception Challenge (Workshop on Autonomous Driving at CVPR 2021) using a single YOLOX-L model. We hope this report can provide useful experience for developers and researchers in practical scenes, and we also provide deploy versions with ONNX, TensorRT, NCNN, and Openvino supported.
+
+
+
+
+

+
+

+
 +
+ +
+ +
+ +
+