+
+
+
+
+User Story 3 - From SSRs to pan genomes with Pretzel: Two decades of wheat yield QTLs on chromosome 7A
+There are three main parts to this User Story:
+
+-
+
In the first part, we will learn how to navigate the wheat pan genome in Pretzel, and visualise some well known results such as the 5B/7B translocation in ArinaLrFor and SY Mattis as well as the Thinopyrum ponticum introgression in LongReach Lancer at the end of 3DL.
+
+-
+
In the second part, we will briefly summarise results from the last 20 years around yield on chromosome 7A. We will use Pretzel to integrate a range of QTL studies and ultimately identify the WAPO1 gene.
+
+-
+
Finally, we will combine the first two parts to:
+
+- Reproduce results reported in Kuzay et al. 2019 and Voss-Fels et al. 2019 relating to haplotypes carried by the 10+ Wheat Genomes in the WAPO1 region of chromosome 7A;
+- Extend these results to classify wheat PGR accessions in the AGG;
+- Identify wheat PGR accessions in the AGG carrying rare haplotypes in the WAPO1 region.
+
+
+
+Details for all publications cited are listed in the references.
+Login
+Log in at https://agg.plantinformatics.io/login using the details below:
+Email Address (username)
+UserStory3@AGG
+
+Password
+UserStory3
+
+
+
+
Note
+
Please use the provided login account so you have access to all the relevant data
+
After logging in, click Map Viewer in the top left to enter the Pretzel application.
+
+Step by step with screenshots
+Assemblies from the 10+ Wheat Genome project in Pretzel
+The 10 chromosome-scale assemblies from Walkowiak et al. 2020 have been curated and made available in AGG Pretzel: ArinaLrFor, CDC Landmark, CDC Stanley, Jagger, Julius, LongReach Lancer, Mace, Norin 61, SY Mattis and spelt wheat PI 190962.
+
+Metadata about the assemblies can be viewed by clicking on the respective Genome entry in the Pretzel Datasets Explorer. The metadata has been extracted from Walkowiak et al. 2020 and includes accession name, pedigree, growth habit, origin and EBI-ENA ID.
+
+Gene annotations by PGSB
+The latest de novo gene annotations from White et al. 2024 (https://doi.org/10.1101/2024.01.09.574802) have been loaded into Pretzel.
+Example: Viewing a genome annotation
+For example, we can load the PGSB annotation for CDC Landmark chromosome 2B by selecting chr2B from the Triticum aestivum - CDC Landmark - Genes PGSBv2.1 dataset. We can click the axis title to open the axis title menu, split the axis, zoom in and select a region. The selected genes are shown in the Features tab in the right panel.
+
+Infinium Wheat Barley 40K v1.1 SNP array marker mappings
+Markers from the Infinium Wheat Barley 40K v1.1 SNP array, which is being used to genotype the AGG wheat collection, have been mapped to each of the 10+ Wheat genomes using Brioche.
+The mapping of markers across the genomes enables chromosome-scale alignments to be visualised, and regions to be projected from one genome to another (similar to how regions were projected between IWGSC RefSeq v1.0 and IWGSC RefSeq v2.1 in User Story 2).
+Example: Visualising the 5B/7B translocation in ArinaLrFor and SY Mattis
+Walkowiak et al. 2020 reported a striking translocation between 5B and 7B found in ArinaLrFor and SY Mattis. We can visualise this by loading the Wheat Barley 40K marker mapping in IWGSC RefSeq v2.1 for chromosomes 5B and 7B, and the same chromosomes in ArinaLrFor and/or SY Mattis.
+
+We can clearly see the long arm of 7B has joined with the long arm of 5B in ArinaLrFor. Rearranging the order of the chromosomes, we can also see how the short arm of 5B has joined to the short arm of 7B in ArinaLrFor. To re-order the axes, hold Ctrl then click on the axis and drag it.
+
+Example: Identifying the genes underlying the Thinopyrum ponticum introgression in LongReach Lancer at the end of 3DL
+First, we load the Wheat Barley 40K marker mapping for chromosome 3D in both IWGSC RefSeq v2.1 and LongReach Lancer.
+
+Note that the LongReach Lancer chromosome 3D is longer than the Chinese Spring 3D.
+Next, we can load the PGSB gene annotation for LongReach Lancer chromosome 3D, split the axis to be able to view the features, then zoom in to the region. We can see a list of the genes by selecting the region and inspecting the Features tab in the right panel. We can reduce the threshold Pretzel uses to display features as histograms by increasing the Threshold slider in the Selected Axis Options section of the View tab in the left panel until features are displayed. Once features are displayed, they will be listed in the Features table when selected.
+
+BLAST search
+Each of the 10+ Wheat genome assemblies can be searched by nucleotide sequence using Pretzel's BLAST search feature. Note that currently, searches can only be done against a single assembly at a time.
+A history of wheat yield QTLs on chromosome 7AL
+Yield and yield-related QTLs have been reported on 7AL since at least 2006. Quarrie et al. 2006 reported a QTL in the centromere (Qyld.csdh.7AC) and on 7AL (Qyld.csdh.7AL), defined in an SSR-based genetic map from SQ1 x Chinese Spring.
+The SSR map for 7AL and SSR marker positions against IWGSC RefSeq v2.1 have been curated and loaded into Pretzel as User Story 3 - Wheat genetic map Chinese Spring x SQ1 and User Story 3 - Wheat SSRs anchored to IWGSC RefSeq v2.1 respectively. Viewing both of these datasets visualises the genetic to physical alignment of this map.
+
+Ten years later, Su et al. 2016 reported a QTL for Thousand Kernel Weight (TKW) on 7AL. Keeble-Gagnère et al. 2018 also reported a similar yield QTL from a RAC785 x Kukri cross.
+In 2019, two studies (Kuzay et al. 2019 and Voss-Fels et al. 2019) reported QTLs for SNS and NRN in the same region.
+This set of QTLs have been curated in the dataset User Story 3 - Wheat yield QTLs on 7A, defined as intervals against IWGSC RefSeq v2.1, based on curation of the marker intervals reported in the papers.
+In Pretzel we can integrate all these results together and see how the interval was progressively narrowed. Ultimately, both Kuzay et al. 2019 and Voss-Fels et al. 2019 identified the wheat ortholog of rice gene APO (ABERRANT PANICLE ORGANIZATION), named WAPO1, as the likely gene influencing the yield phenotype.
+First, load the User Story 3 - Wheat yield QTLs on 7A dataset. Then switch to the View tab, and under the Displayed Data section at the top, click the Trait tab. Then tick the box titled Show / Hide QTLs of all Traits.
+
+A brief detour into wheat-rice synteny, or, how we found the WAPO1 gene
+Note: This brief section can only be reproduced on https://plantinformatics.io for now. Datasets from plantinformatics.io will be transfered to AGG Pretzel over the next 6 months. Those interested in studying wheat-rice syntenic alignments can sign up for a free account at https://plantinformatics.io/signup.
+Sign in to https://plantinformatics.io and load the following datasets: chromosomes 6 and 8 from Oryza_sativa_IRGSP-1.0_genes and chr7A from Triticum_aestivum_IWGSC_RefSeq_v1.0_HC_genes. Re-order the axes so that 7A is in the middle.
+
+We have visualised the striking syntenic relationship between the wheat group 7 chromosomes and rice chromosomes 6 and 8. Next, we can search for the rice ABERRANT PANICLE ORGANIZATION (APO) gene ID, Os06g0665400. The search results identify the rice gene but also the orthologous wheat ID, TraesCS7A01G481600.
+
+Confirming the location of the WAPO1 gene
+We can use the WAPO1 gene sequence to locate the gene in the genome. Copy the sequence from the BLAST Use Case and follow the steps in that Use Case to search the IWGSC RefSeq v2.1 assembly. If searching with default parameters (as in the animation below), the BLAST results identify the 7A, 7B and 7D homoeologs of the gene. Remove the 7B and 7D chromosomes from the view.
+
+We can now zoom closer to the region to find that the gene falls exactly within the set of overlapping intervals, but outside the interval from Quarrie et al. 2006.
+
+Before we proceed, we will remove the SSR map from the view, split the IWGSC RefSeq v2.1 chromosome 7A axis and adjust the view to capture the complete region of interest.
+
+Note that we can view details about the QTLs by selecting the region and inspecting the Features tab in the right panel.
+
+Studying the WAPO1 region in the 10+ Wheat genomes
+A key result from Kuzay et al. 2019 and Voss-Fels et al. 2019 was that only two major haplotypes in the WAPO1 region of 7A, named HAP1 and HAP2, dominate modern hexaploid wheat, with HAP2 associated with increased grain yield. Kuzay et al. 2019 classified the haplotypes of the 10+ Wheat genomes and found that ArinaLrFor, CDC Landmark, Jagger, Julius, LongReach Lancer and SY Mattis carry HAP1 while Chinese Spring, CDC Stanley, Mace and Norin61 carry HAP2. The spelt accession included in the 10+ Wheat genomes carried a different haplotype.
+Reproducing the haplotype classification from Kuzay et al. 2019
+Using Brioche, a tool developed in the Australian Grains Genebank Strategic Partnership, we have in-silico genotyped the 10+ Wheat genome assemblies with the Wheat Barley 40K v1.1 SNP array. This dataset is available on AGG Pretzel as Triticum aestivum - IWGSC RefSeq v2.1 - Genotypes - 10 Wheat Genomes.
+Continuing from the above analysis where we have located the WAPO1 region, we can load this dataset, select the region around the gene, and load genotypes for the 10+ Wheat genomes. Clicking the ALT column, we can sort the accessions based on their allele at the selected SNP.
+
+We have reproduced the same haplotype classification as reported in Kuzay et al. 2019. This shows that the Wheat Barley 40K v1.1 SNP array is able to correctly differentiate the haplotypes reported in the paper, noting that exome sequence was used in the 2019 study.
+Connecting the pan genome to the AGG
+We have so far identified the WAPO1 region on chromosome 7AL through the integration of a number of yield QTLs over almost 20 years of research. We have reproduced the haplotype analysis reported in Kuzay et al. 2019, confirming the Wheat Barley 40K v1.1 SNP array detects the main haplotypes found in modern wheat.
+We can now use Pretzel to directly relate these results to the AGG. We will achieve two main things: 1) Confirm that most wheat accessions in the AGG carry one of the two dominant haplotypes (HAP1 and HAP2) in the WAPO1 region; 2) Identify rarer haplotypes carried by PGR accessions in the AGG.
+Continuing from the previous step where we visualised genotypes for the 10 Wheat genomes, we will now bring in genotypes for wheat PGR accessions in the AGG. In the Dataset Explorer, add chromosome 7A from the Triticum aestivum - IWGSC_RefSeq_v2.1 - Genotypes - AGG Filled-in Release 1 dataset. In the Genotypes menu there are now two tabs - the first - Wheat_pangenomes_IWGSC_v2.1 - is the 10 Wheat genomes, the second - Wheat_CSv2.1_VCF-AGG-imp-r1 - is the AGG data. Select the second tab then select some accessions from the list. To select multiple accessions in a row, click on the first accession in the list, then while holding Shift, click an accession further down the list to select all accessions in between. In this example we select the first 150 or so accessions. If you have accessions of interest, you could search for them and add those to the list. Clicking VCF Lookup will request the genotype data for these accessions and add them to the genotype table.
+Clicking the ALT allele for each of the 5 SNPs that make up the haplotype will order accessions based on their distance from HAP1 (all 5 ALT alleles). Scrolling to the right we can see most of the AGG accessions carry either HAP1 or HAP2 (all REF alleles). In between we can clearly see accessions carrying rarer haplotypes, including the haplotype carried by the spelt accession. We have achieved our initial aim of finding accessions in the AGG carrying rare haplotypes in the WAPO1 region.
+
+References
+International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018 Aug 17;361(6403):eaar7191. doi: 10.1126/science.aar7191. Epub 2018 Aug 16. PMID: 30115783.
+Keeble-Gagnère G, Rigault P, Tibbits J, Pasam R, Hayden M, Forrest K, Frenkel Z, Korol A, Huang BE, Cavanagh C, Taylor J, Abrouk M, Sharpe A, Konkin D, Sourdille P, Darrier B, Choulet F, Bernard A, Rochfort S, Dimech A, Watson-Haigh N, Baumann U, Eckermann P, Fleury D, Juhasz A, Boisvert S, Nolin MA, Doležel J, Šimková H, Toegelová H, Šafář J, Luo MC, Câmara F, Pfeifer M, Isdale D, Nyström-Persson J, Iwgsc, Koo DH, Tinning M, Cui D, Ru Z, Appels R. Optical and physical mapping with local finishing enables megabase-scale resolution of agronomically important regions in the wheat genome. Genome Biol. 2018 Aug 17;19(1):112. doi: 10.1186/s13059-018-1475-4. PMID: 30115128; PMCID: PMC6097218.
+Kuzay S, Xu Y, Zhang J, Katz A, Pearce S, Su Z, Fraser M, Anderson JA, Brown-Guedira G, DeWitt N, Peters Haugrud A, Faris JD, Akhunov E, Bai G, Dubcovsky J. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor Appl Genet. 2019 Sep;132(9):2689-2705. doi: 10.1007/s00122-019-03382-5. Epub 2019 Jun 28. PMID: 31254024; PMCID: PMC6708044.
+Quarrie S, Pekic Quarrie S, Radosevic R, Rancic D, Kaminska A, Barnes JD, Leverington M, Ceoloni C, Dodig D. Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot. 2006;57(11):2627-37. doi: 10.1093/jxb/erl026. Epub 2006 Jul 10. PMID: 16831847.
+Su, Z., Jin, S., Lu, Y. et al. Single nucleotide polymorphism tightly linked to a major QTL on chromosome 7A for both kernel length and kernel weight in wheat. Mol Breeding 36, 15 (2016). https://doi.org/10.1007/s11032-016-0436-4
+Voss-Fels KP, Keeble-Gagnère G, Hickey LT, Tibbits J, Nagornyy S, Hayden MJ, Pasam RK, Kant S, Friedt W, Snowdon RJ, Appels R, Wittkop B. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theor Appl Genet. 2019 Sep;132(9):2707-2719. doi: 10.1007/s00122-019-03383-4. Epub 2019 Jun 28. PMID: 31254025.
+Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, Klymiuk V, Byrns B, Gundlach H, Bandi V, Siri JN, Nilsen K, Aquino C, Himmelbach A, Copetti D, Ban T, Venturini L, Bevan M, Clavijo B, Koo DH, Ens J, Wiebe K, N'Diaye A, Fritz AK, Gutwin C, Fiebig A, Fosker C, Fu BX, Accinelli GG, Gardner KA, Fradgley N, Gutierrez-Gonzalez J, Halstead-Nussloch G, Hatakeyama M, Koh CS, Deek J, Costamagna AC, Fobert P, Heavens D, Kanamori H, Kawaura K, Kobayashi F, Krasileva K, Kuo T, McKenzie N, Murata K, Nabeka Y, Paape T, Padmarasu S, Percival-Alwyn L, Kagale S, Scholz U, Sese J, Juliana P, Singh R, Shimizu-Inatsugi R, Swarbreck D, Cockram J, Budak H, Tameshige T, Tanaka T, Tsuji H, Wright J, Wu J, Steuernagel B, Small I, Cloutier S, Keeble-Gagnère G, Muehlbauer G, Tibbets J, Nasuda S, Melonek J, Hucl PJ, Sharpe AG, Clark M, Legg E, Bharti A, Langridge P, Hall A, Uauy C, Mascher M, Krattinger SG, Handa H, Shimizu KK, Distelfeld A, Chalmers K, Keller B, Mayer KFX, Poland J, Stein N, McCartney CA, Spannagl M, Wicker T, Pozniak CJ. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020 Dec;588(7837):277-283. doi: 10.1038/s41586-020-2961-x. Epub 2020 Nov 25. PMID: 33239791; PMCID: PMC7759465.
+Benjamen White, Thomas Lux, Rachel Rusholme-Pilcher, Angéla Juhász, Gemy Kaithakottil, Susan Duncan, James Simmonds, Hannah Rees, Jonathan Wright, Josh Colmer, Sabrina Ward, Ryan Joynson, Benedict Coombes, Naomi Irish, Suzanne Henderson, Tom Barker, Helen Chapman, Leah Catchpole, Karim Gharbi, Utpal Bose, Moeko Okada, Hirokazu Handa, Shuhei Nasuda, Kentaro K. Shimizu, Heidrun Gundlach, Daniel Lang, Guy Naamati, Erik J. Legg, Arvind K. Bharti, Michelle L. Colgrave, Wilfried Haerty, Cristobal Uauy, David Swarbreck, Philippa Borrill, Jesse A. Poland, Simon G. Krattinger, Nils Stein, Klaus F.X. Mayer, Curtis Pozniak, 10+ Wheat Genome Project, Manuel Spannagl, Anthony Hall. De novo annotation of the wheat pan-genome reveals complexity and diversity of the hexaploid wheat pan-transcriptome. bioRxiv 2024.01.09.574802; doi: https://doi.org/10.1101/2024.01.09.574802
+Zhu T, Wang L, Rimbert H, Rodriguez JC, Deal KR, De Oliveira R, Choulet F, Keeble-Gagnère G, Tibbits J, Rogers J, Eversole K, Appels R, Gu YQ, Mascher M, Dvorak J, Luo MC. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021 Jul;107(1):303-314. doi: 10.1111/tpj.15289. Epub 2021 May 16. PMID: 33893684; PMCID: PMC8360199.
+
+
+
+
+
+
+
+
+
+
+
+
+
+ 
 +Within the 'VCF Genotype Search' box, select the drop down menu under 'VCF to search :' select
+Within the 'VCF Genotype Search' box, select the drop down menu under 'VCF to search :' select  +Witin the same pannel input the following into 'Samples input :'
+Witin the same pannel input the following into 'Samples input :'
 +2. Clicking on the name of a dataset will highlight it and show information
+about the data set on the right hand pannel
+2. Clicking on the name of a dataset will highlight it and show information
+about the data set on the right hand pannel +3. Pressing the plus button will expand the dataset to reveal items that can be loaded into the view
+3. Pressing the plus button will expand the dataset to reveal items that can be loaded into the view  +4. Pressing the green plus buttons will then bring that item into the view
+4. Pressing the green plus buttons will then bring that item into the view +5. If the item is no longer need it can be removed by clicking on the title of the item in the view to reveal an additional menu. Then select the cross button in the centre
+5. If the item is no longer need it can be removed by clicking on the title of the item in the view to reveal an additional menu. Then select the cross button in the centre +6. The dataset is now removed from view
+6. The dataset is now removed from view
 +2. The results of the search will up down in the bottom most area coloured in blue. With the number just above the number of returned results.
+2. The results of the search will up down in the bottom most area coloured in blue. With the number just above the number of returned results.






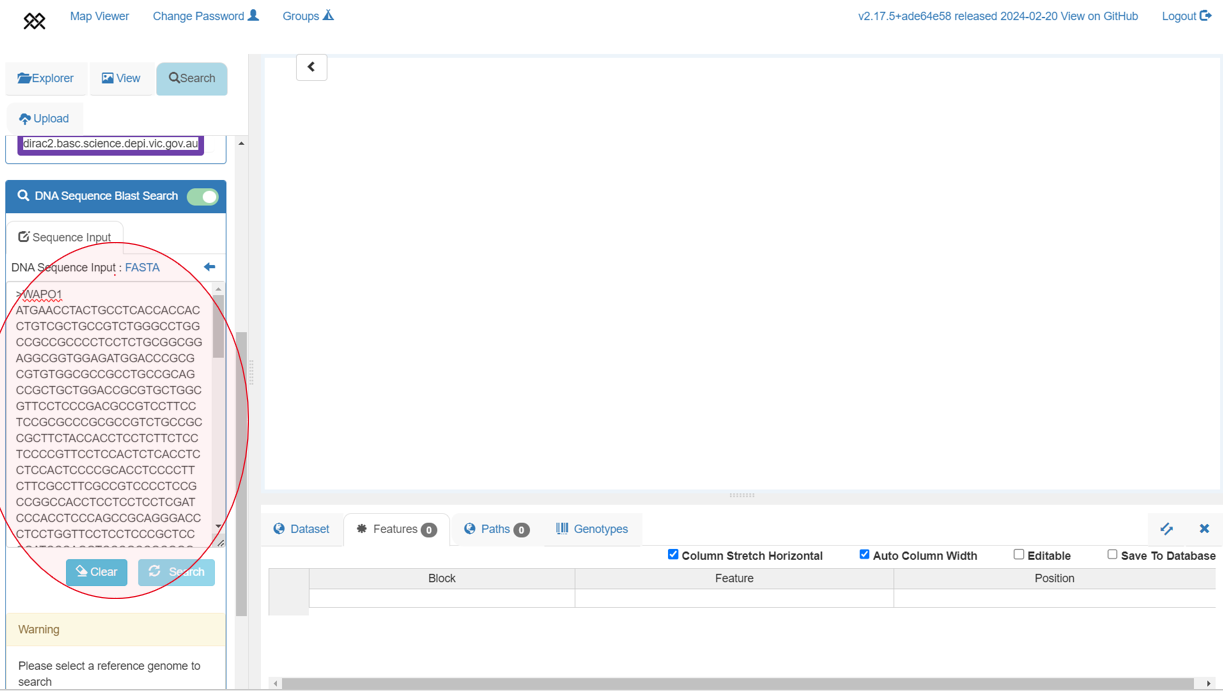
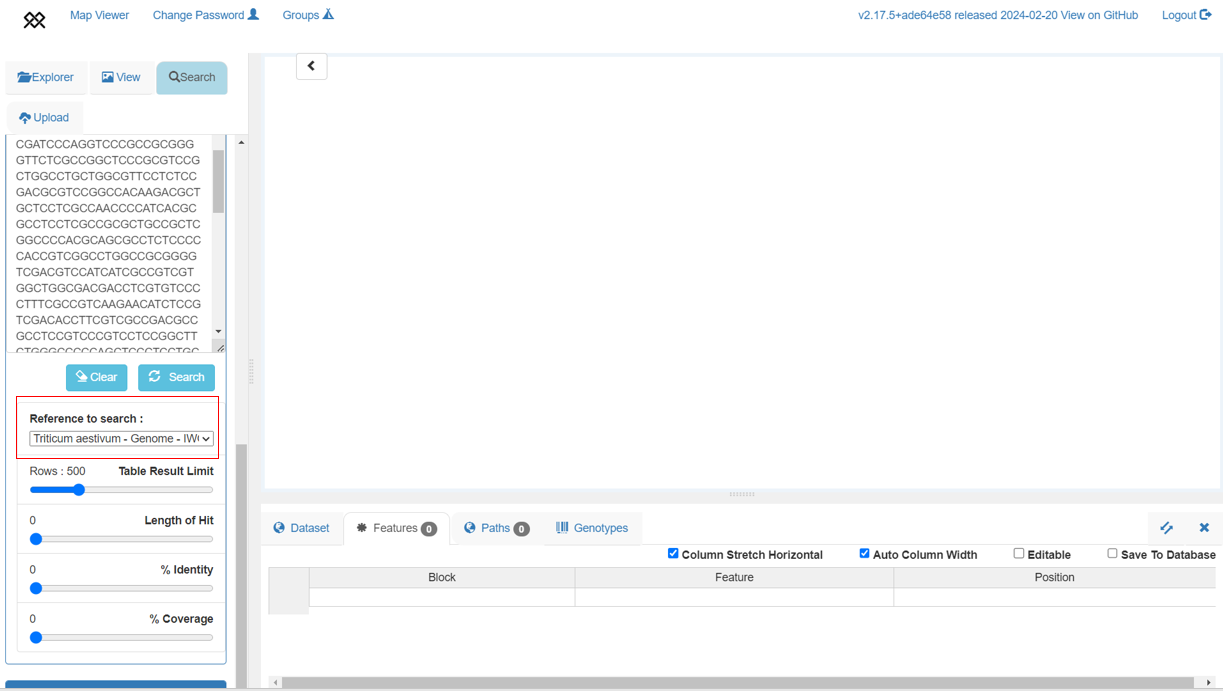
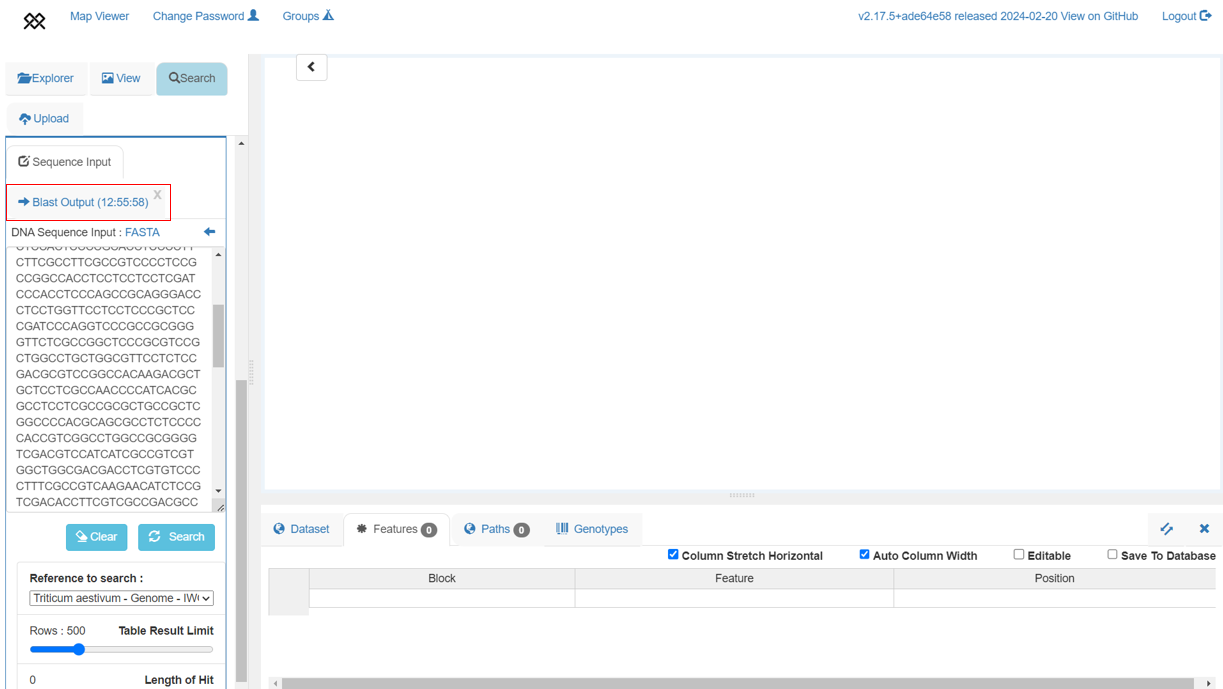



















 +To close this box press the top right "x" button
+
+To close this box press the top right "x" button
+
 +An arrow should appear on left of the axis and click the arrow to bring the up name
+
+An arrow should appear on left of the axis and click the arrow to bring the up name
+






















