-
Notifications
You must be signed in to change notification settings - Fork 5
Walkthrough data
This wiki page provides an overview of how to import new data sets.
The scenario is that we have just been given a new raw EEG file, that we want to fit using BrainTrak. In this case, we have recieved 2 .raw files.
The first thing to do is to establish the folder structure, thus naming the data set. In this case, the data corresponds to subjects in an unresponsive/vegetative state, so we can create a folder called unresponsive and put the raw EEG files in there. It is also helpful to use separate folders for different subject groups. For example, a corresponding control data set might be in a folder unresponsive_control to reduce the possibility of mixing up the two data sets.
Next, the files should be renamed to a standard numbering scheme. For example, the two files recieved were 20101104.raw and 20140522.raw. These should be renamed to unresponsive_1.raw and unresponsive_2.raw. Make sure you save a text file (maybe a CSV if you like) recording this. For example, mapping.csv could contain
20140522.raw,unresponsive_1.raw
20140522.raw,unresponsive_2.raw
Keep this file together with the data files.
Next, we have been told that these files are "256 EEG from EGI". We need to convert the .raw files into .mat files. In general, reading the binary files correctly is difficult to do, so wherever possible, you should use existing code to perform the conversion. A good place to start is EEGLAB which has plugins to read most common file formats. The documentation in EEGLAB shows that EGI/RAW files can be loaded using the Biosig extension, so we can start EEGLAB and go to File > Import data > Using the Biosig interface and select unresponsive_1.raw.
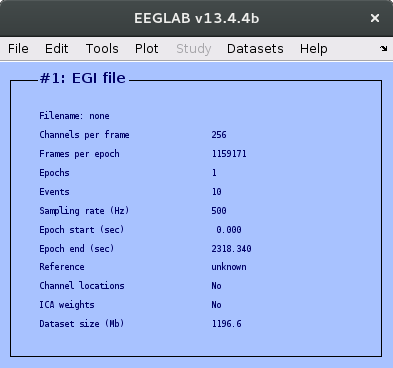
Oops, that didn't work! This is to be expected when working with raw data files. More google searching suggests that the EGI raw files are the same as Netstation simple binary files, so we can use File > Import data > Using EEGLAB functions and plugins > From Netstation binary simple file. Following the prompts and giving a sensible name for the data set leads to the file being successfully loaded. We can verify that the number of channels is correct, and that the sampling rate and epoch start and end look sensible.
Looking at bt.data.import_raw_eeg, we can see what variables are required. We need to have the following variables:
-
colheaderscorresponding to the electrode names -
tcorresponding to the time values -
datacorresponding to a matrix where the rows are time, and the columns are electrodes -
epoch_scorewhich corresponds to the sleep stage
EEGLAB loads the data file into the workspace variable ALLEEG. We can extract parts of ALLEEG to form the above variables.
colheaders = {ALLEEG.urchanlocs.labels}; % Note conversion to cell array
t = ALLEEG.times(:); % Note this is being converted to a column vector
data = ALLEEG.data.'; % Pay attention to the row/column orientation
For epoch_score, this is not a sleep data set, so we should just pick a sensible value. We can add a row to the matrix in bt_utils.state_cdata and use the corresponding index for our new dataset, to give it a different colour. For example, the repository has row 8 corresponding to the 'unresponsive' state, so we can set
epoch_score = 8*ones(size(t));
Lastly, we can save our converted file. For example,
save unresponsive/unresponsive_1.mat colheaders t data epoch_score
We now repeat this process for all of the other files. Note that EEGLAB by default will append the new file to ALLEEG. You need to first select File > Clear study / Clear all before loading the next file.
Note that our strategy has been to do as little processing to the raw data as possible, in converting the raw data to .mat format. Thus unresponsive_1.mat can be used as the starting point for most subsequent analyses. This has the advantage of making it very unlikely that you will need to revisit the .raw file. Now we're done with EEGLAB and can start working with BrainTrak.
All of the electrode potentials are measured relative to something - typically we reference to A1+A2, or to A1 or A2 on the opposite side of the head. Alternatively, the average of all electrodes can be used, which may be preferable in data sets like this. You can do the re-referencing yourself by subtracting channels from each other, or you can do this from within EEGLAB. You should check with your collaborator what the existing reference is. You should state in any papers you write what reference you used.
The raw data file now needs to be converted into a format suitable for BrainTrak. We need to make appropriate adaptation to bt.data.import_raw_eeg to perform the conversion. It's a good idea to make a copy of this file, and put it somewhere in your project directory or the data_examples folder. So here, we will make data_examples/import_unresponsive.m.
For development, it can be useful for debugging to set idx = 1 or similar (only work with one file) and to change the parfor loop to a for loop to aid debugging.
You will need to change the input file location - for example,
infile = fullfile(os_prefix,'unresponsive',sprintf('%s_%d.mat',data_set,k));
Also, around line 52 where the output file is being saved, you should change the file location. By convention, BrainTrak .mat files end with _tfs.mat to identify that they contain the t, f and s variables generated by bt.data.get_tfs. For example,
outfile = fullfile(os_prefix,'unresponsive',sprintf('%s_%d_tfs.mat',data_set,k));
Running import_unresponsive now prints diagnostic output while importing the electrodes
>> import_unresponsive
Loading: ./psg_data/unresponsive/unresponsive_1.mat
unresponsive_1: E1 electrode
...
Now we get an error! Diagnosing the error reveals that the file saved from EEGLAB has a time vector that looks like
t = [0 2 4 ...]
This does not correspond to seconds elapsed in the recording. So we need to reconstruct the time array. We know that the data has a sampling rate of 500 Hz (this was shown in EEGLAB earlier) so we can repair the data files as follows:
clear all
load ./psg_data/unresponsive/unresponsive_1.mat
fs = 500;
t_end = ceil(length(t)/fs); % What is the end time?
t = 0:1/fs:t_end;
t = t(1:size(data,1)).'; % Check that the sizes match the original arrays
save ./psg_data/unresponsive/unresponsive_1.mat colheaders t data epoch_score
As a general rule, 1/t(2) should be equal to the sampling rate, and t needs to have the same number of rows as data. The same fix can be applied to the other files. Of course, it would have been better to have checked this earlier when saving the initial files from EEGLAB.
Now we can try import_unresponsive again, and get_tfs works, but the sleep stages aren't handled properly. We need to set
-
state_str, which is a string describing the brain state e.g. EC -
state_scorewhich is an index ofbt_utils.state_cdata
We can copy epoch_score straight into state_score and set state_str to be 'Unresponsive' at all times:
state_str = cell(length(t),1);
[state_str{:}] = deal('Unresponsive');
state_score = fhandle.epoch_score;
And we can strip out the existing code for assigning these variables. Finally, we can run import_unresponsive and have the conversion run to completion. For each file, our data directory now contains
-
unresponsive_1.raw- EEG time series data from recording system -
unresponsive_1.mat- EEG time series data in.matformat -
unresponsive_1_tfs.mat- EEG power spectrum suitable for BrainTrak
Many BrainTrak scripts load data via bt.core.load_subject. To add support for our data set, we need to add an appropriate statement to the if block. For example, we could have
elseif any(strcmp({'unresponsive'},dataset))
fdata = load(sprintf('./psg_data/unresponsive/%s_%d_tfs',dataset,subject_idx));
We also need to select a default electrode. This data set has 256 channels with names like E200. By convention, load_subject should provide a default electrode close to Cz. Searching for 'egi electrode coordinates' leads to this information page that points to a PDF file that maps EGI electrodes to the standard names in the 10-10 system. If we decided that E161 is the electrode we wanted, we can add
elseif strcmp(dataset,'unresponsive')
electrode = {'E161'};
The proper choice of electrode is something that should be discussed with the collaborator providing the data.
You can now test that the spectrum can be loaded and plotted correctly.
d = bt.core.load_subject('unresponsive',1)
loglog(d.f,d.s(:,1))
And then fit the spectrum using bt.fit_track:
f = bt.fit_track(bt.model.full_emgf,'unresponsive',1,1:10)
f.plot()
