This repository contains the code for the OpenPredict Translator API available at openpredict.semanticscience.org, which serves a few prediction models developed at the Institute of Data Science.
- various folders for different prediction models served by the OpenPredict API are available under
src/:- the OpenPredict drug-disease prediction model in
src/openpredict_model/ - a model to compile the evidence path between a drug and a disease explaining the predictions of the OpenPredict model in
src/openpredict_evidence_path/ - a prediction model trained from the Drug Repurposing Knowledge Graph (aka. DRKG) in
src/drkg_model/
- the OpenPredict drug-disease prediction model in
- the code for the OpenPredict API endpoints in
src/trapi/defines:- a TRAPI endpoint returning predictions for the loaded models
The data used by the models in this repository is versionned using dvc in the data/ folder, and stored on DagsHub at https://dagshub.com/vemonet/translator-openpredict
Requirements: Python 3.8+ and pip installed
-
Clone the repository with submodule:
git clone --recursive https://github.com/MaastrichtU-IDS/translator-openpredict.git cd translator-openpredict -
Pull the data required to run the models in the
datafolder withdvc:pip install dvc dvc pull
Start the API in development mode with docker on http://localhost:8808, the API will automatically reload when you make changes in the code:
docker compose up apiContributions are welcome! If you wish to help improve OpenPredict, see the instructions to contribute 👩💻 for more details on the development workflow
Run the tests locally with docker:
docker compose run testsSee the
TESTING.mdfile for more details on testing the API.
You can change the entrypoint of the test container to run other commands, such as training a model:
docker compose run --entrypoint "python src/openpredict_model/train.py train-model" tests
# Or with the helper script:
./resources/run.sh python src/openpredict_model/train.py train-modelThe user provides a drug or a disease identifier as a CURIE (e.g. DRUGBANK:DB00394, or OMIM:246300), and choose a prediction model (only the Predict OMIM-DrugBank classifier is currently implemented).
The API will return predicted targets for the given drug or disease:
- The potential drugs treating a given disease 💊
- The potential diseases a given drug could treat 🦠
Feel free to try the API at openpredict.semanticscience.org
Operations to query OpenPredict using the Translator Reasoner API standards.
The /query operation will return the same predictions as the /predict operation, using the ReasonerAPI format, used within the Translator project.
The user sends a ReasonerAPI query asking for the predicted targets given: a source, and the relation to predict. The query is a graph with nodes and edges defined in JSON, and uses classes from the BioLink model.
You can use the default TRAPI query of OpenPredict /query operation to try a working example.
Example of TRAPI query to retrieve drugs similar to a specific drug:
{
"message": {
"query_graph": {
"edges": {
"e01": {
"object": "n1",
"predicates": [
"biolink:similar_to"
],
"subject": "n0"
}
},
"nodes": {
"n0": {
"categories": [
"biolink:Drug"
],
"ids": [
"DRUGBANK:DB00394"
]
},
"n1": {
"categories": [
"biolink:Drug"
]
}
}
}
},
"query_options": {
"n_results": 3
}
}The /predicates operation will return the entities and relations provided by this API in a JSON object (following the ReasonerAPI specifications).
Try it at https://openpredict.semanticscience.org/predicates
We provide Jupyter Notebooks with examples to use the OpenPredict API:
- Query the OpenPredict API
- Generate embeddings with pyRDF2Vec, and import them in the OpenPredict API
The default baseline model is openpredict_baseline. You can choose the base model when you post a new embeddings using the /embeddings call. Then the OpenPredict API will:
- add embeddings to the provided model
- train the model with the new embeddings
- store the features and model using a unique ID for the run (e.g.
7621843c-1f5f-11eb-85ae-48a472db7414)
Once the embedding has been added you can find the existing models previously generated (including openpredict_baseline), and use them as base model when you ask the model for prediction or add new embeddings.
Use this operation if you just want to easily retrieve predictions for a given entity. The /predict operation takes 4 parameters (1 required):
- A
drug_idto get predicted diseases it could treat (e.g.DRUGBANK:DB00394)- OR a
disease_idto get predicted drugs it could be treated with (e.g.OMIM:246300)
- OR a
- The prediction model to use (default to
Predict OMIM-DrugBank) - The minimum score of the returned predictions, from 0 to 1 (optional)
- The limit of results to return, starting from the higher score, e.g. 42 (optional)
The API will return the list of predicted target for the given entity, the labels are resolved using the Translator Name Resolver API
Try it at https://openpredict.semanticscience.org/predict?drug_id=DRUGBANK:DB00394
- The gold standard for drug-disease indications has been retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3159979
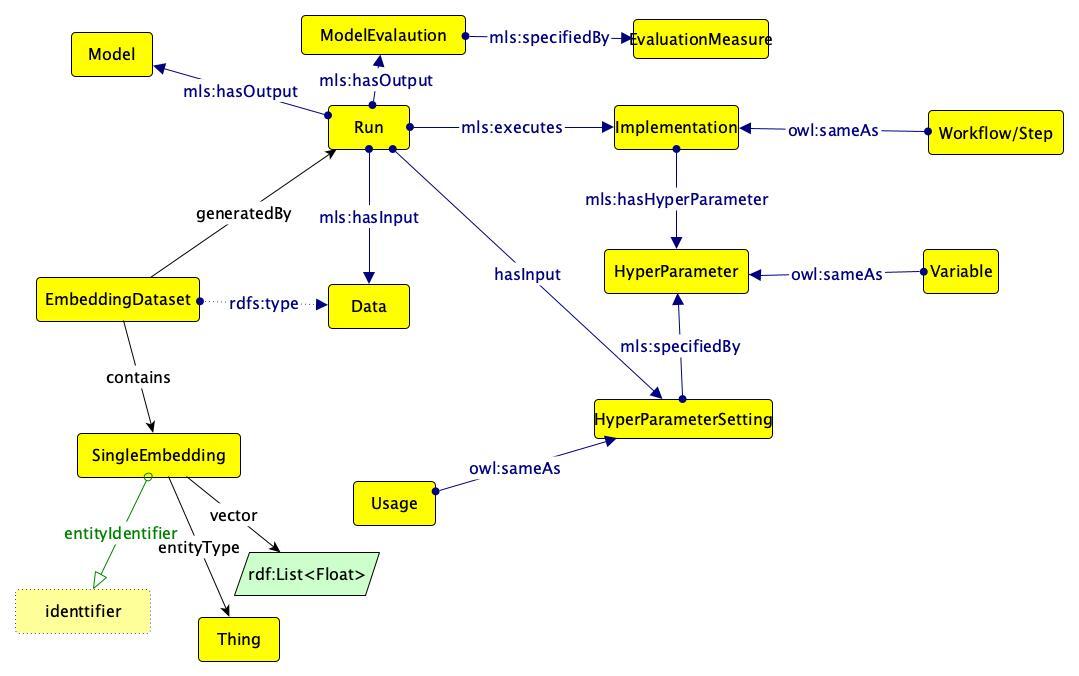
- Metadata about runs, models evaluations, features are stored as RDF using the ML Schema ontology.
- See the ML Schema documentation for more details on the data model.
Diagram of the data model used for OpenPredict, based on the ML Schema ontology (mls):
Query for drug-disease pairs predicted from pre-computed sets of graphs embeddings.
Add new embeddings to improve the predictive models, with versioning and scoring of the models.
API component
-
Component Name: OpenPredict API
-
Component Description: Python API to serve pre-computed set of drug-disease pair predictions from graphs embeddings
-
GitHub Repository URL: https://github.com/MaastrichtU-IDS/translator-openpredict
-
Component Framework: Knowledge Provider
-
System requirements
5.1. Specific OS and version if required: python 3.8
5.2. CPU/Memory (for CI, TEST and PROD): 32 CPUs and 32 Go memory ?
5.3. Disk size/IO throughput (for CI, TEST and PROD): 20 Go ?
5.4. Firewall policies: does the team need access to infrastructure components? The NodeNormalization API https://nodenormalization-sri.renci.org
-
External Dependencies (any components other than current one)
6.1. External storage solution: Models and database are stored in
/data/openpredictin the Docker container -
Docker application:
7.1. Path to the Dockerfile:
Dockerfile7.2. Docker build command:
docker build ghcr.io/maastrichtu-ids/openpredict-api .7.3. Docker run command:
Replace
${PERSISTENT_STORAGE}with the path to persistent storage on host:docker run -d -v ${PERSISTENT_STORAGE}:/data/openpredict -p 8808:8808 ghcr.io/maastrichtu-ids/openpredict-api -
Logs of the application
9.2. Format of the logs: TODO
- This service has been built from the fair-workflows/openpredict project.
- Predictions made using the PREDICT method.
- Service funded by the NIH NCATS Translator project.