The semantic comparisons of Gene Ontology (GO) annotations provide
quantitative ways to compute similarities between genes and gene groups,
and have became important basis for many bioinformatics analysis
approaches. GOSemSim is an R package for semantic similarity
computation among GO terms, sets of GO terms, gene products and gene
clusters. GOSemSim implemented five methods proposed by Resnik,
Schlicker, Jiang, Lin and Wang respectively.
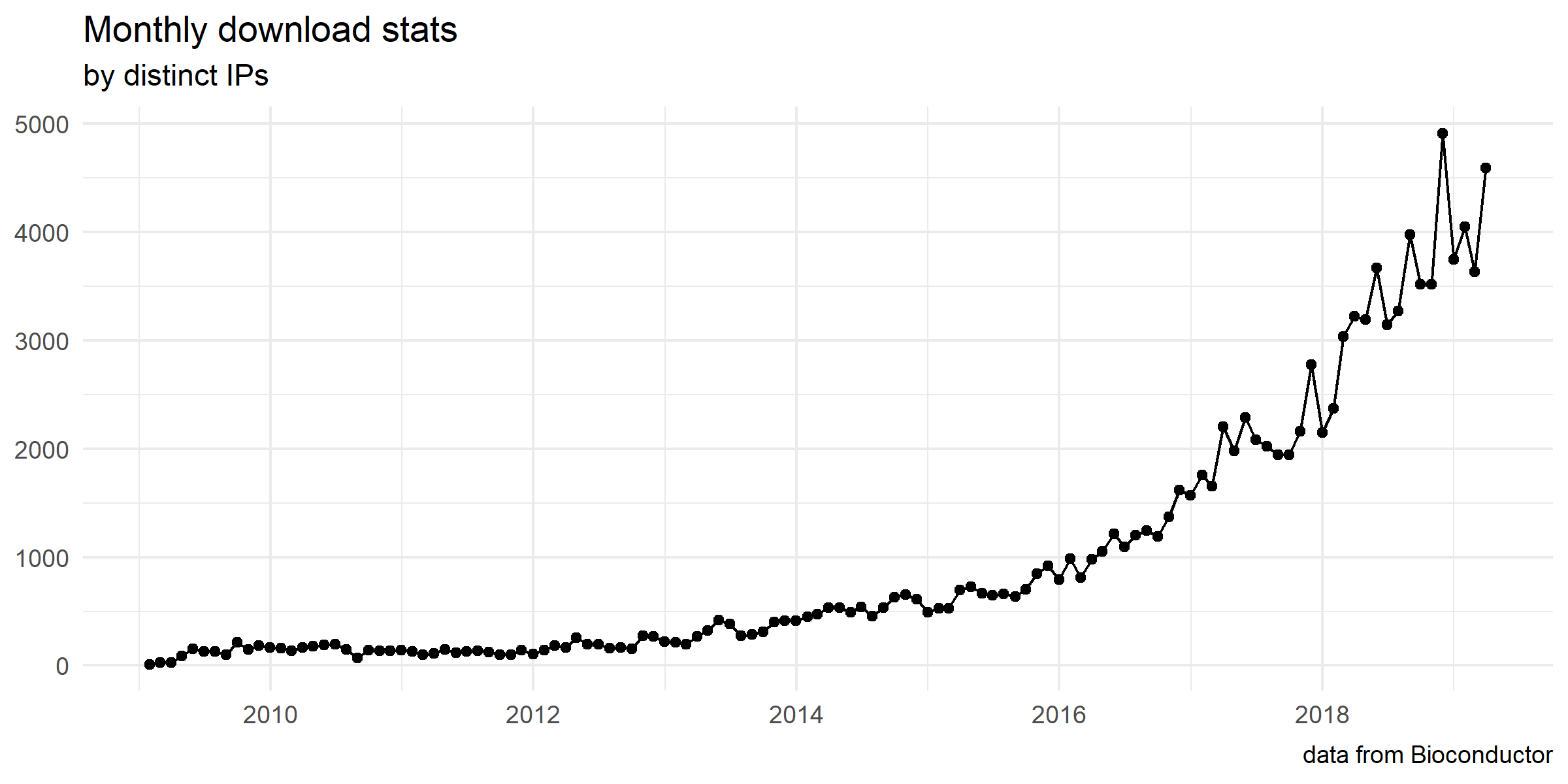
For details, please visit our project website, https://guangchuangyu.github.io/software/GOSemSim.
Please cite the following article when using GOSemSim:
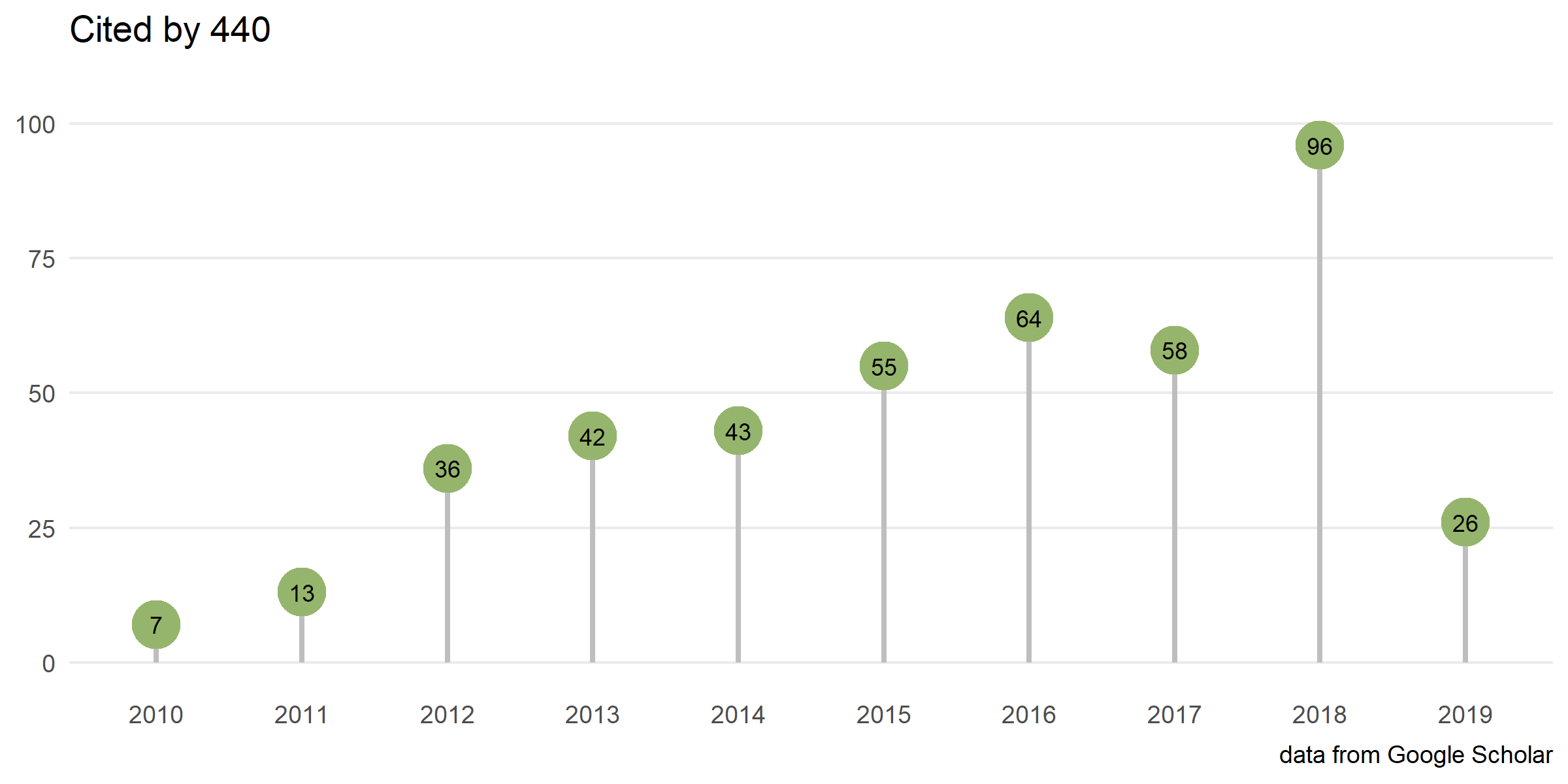
Yu G#, Li F#, Qin Y, Bo X*, Wu Y and Wang S*. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010, 26(7):976-978.
Guangchuang YU https://guangchuangyu.github.io
School of Basic Medical Sciences, Southern Medical University