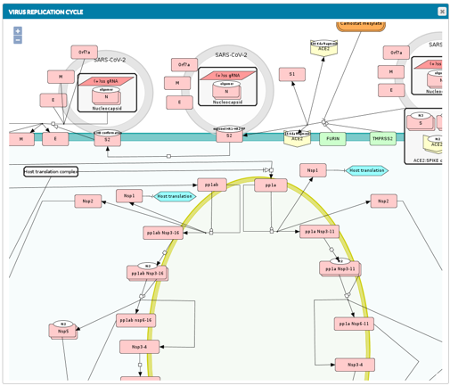
INDRA integrates multiple text-mining systems and pathway databases to automatically extract mechanistic knowledge from the biomedical literature and through a process of knowledge assembly, build executable models and causal networks. Based on profiling and perturbational data, these models can be contextualized to be cell-type specific and used to explain experimental observations or to make predictions.
In the context of the ongoing COVID-19 pandemic, the
INDRA team at the
Laboratory of Systems Pharmacology, Harvard Medical School
is working on understanding the mechanisms by which SARS-CoV-2 infects
cells and the subsequent host response process, with the goal
of finding new therapeutics using INDRA.
EMMAA (Ecosystem of Machine-maintained Models with Automated Analysis) makes available a set of computational models that are kept up-to-date using automated machine reading, knowledge-assembly, and model generation, integrating new discoveries immediately as they become available.
The EMMAA COVID-19 model
integrates all literature made available under the
COVID-19 Open Research Dataset Challenge (CORD-19) and combines it with newly appearing
papers from PubMed (about 300 every day) as well as bioRxiv and
medRxiv preprints. It also integrates content from CTD, DrugBank, VirHostNet,
and many other pathway databases.
- The set of all statements in the model can be browsed and curated
here. - This is a stable link to get the latest dump of all statements in the model
as JSON:
here.
The model is also used to construct casual, mechanistic explanations to around 2,800 drug-virus effects:
- Explanations for drug-virus effects from the
MITRE COVID-19 Therapeutic Information Browseravailablehere. - Explanations for drug-virus effects curated from papers describing
drug-response experiments available
here.
 The EMMAA COVID-19 model is also on Twitter (
The EMMAA COVID-19 model is also on Twitter (@covid19_emmaa) where it provides updates on the findings that it learns from the literature and also
new experimental observations (such as drug effects on viruses, as described
above) that it can explain based on these new pieces of knowledge.
We used INDRA statements assembled from all available biomedical
literature and a multitude of pathway databases to find evidence
for all interactions in the COVID-19 Disease Map, and to suggest other
mechanisms that haven't yet been included. The results are available
here.
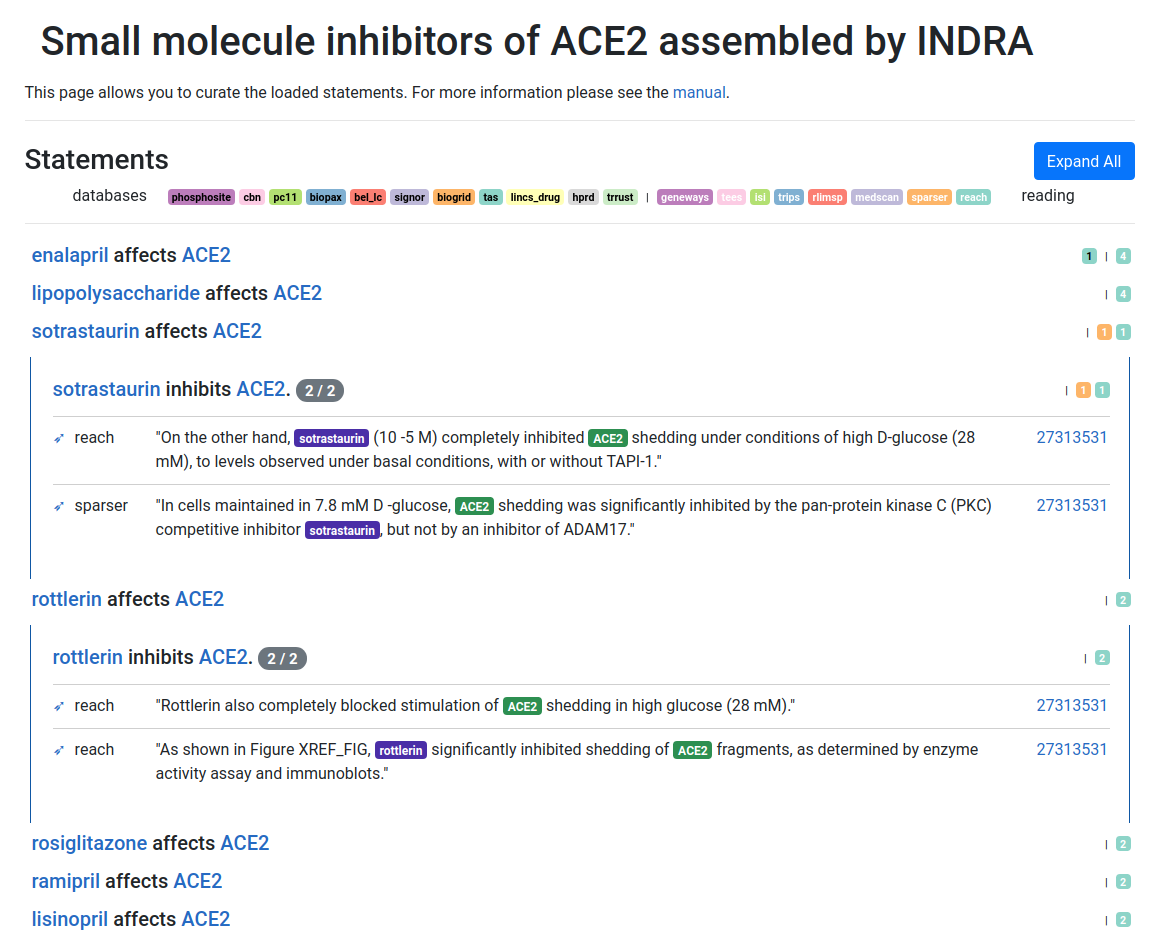
We also implemented a feature - based on the above alignment - to find
small molecule inhibitors for a given pathway in the COVID-19 Disease Map.
The results for the Interferon Type I pathway are available here.
We also used our Gilda system to find
appropriate grounding (database
identifiers) to ungrounded entities used in the Disease Map. The results of
this are available
here.
We also compiled similar reports on the downstream effects of some specific
drugs of interest to our collaborators. These can be found here:
amodiaquine
hydroxychloroquine
While we added some customizations to these reports, similar results can
be obtained by querying the INDRA DB directly.
Another interface for browsing INDRA COVID-19 literature assembly results is available via [`semviz.org`](https://www.semviz.org/) on [`this page`](http://morbius.cs-i.brandeis.edu:23762/login?next=%2Fapp%2Fkibana#/dashboard/2b613e90-7cf0-11ea-8a44-496b85e05ba5) (login: semvizuser/semviz), an approach to semantic browsing of biomedical relations developed at [`Brandeis University`](https://brandeis-llc.github.io/). A tutorial video of using this interface with INDRA results to construct hypotheses about COVID-19 is available [`here`](http://www.voxicon.net/wp-content/uploads/2020/06/semviz.mp4).
CoronaWhy is a globally distributed, volunteer-powered research organisation, assisting the medical community’s ability to answer key questions related to COVID-19.
INDRA is a key part of the CoronaWhy software infrastructure
as an entrypoint to access multiple text-mining systems and pathway databases
and assembling causal models from these sources.
We have developed several applications that are generally applicable to biomedical research and can therefore also be used to study COVID-19.
INDRA: INDRA can be used as aPython packageor aweb serviceto collect relevant information from the literature and pathway databases and build custom COVID-19 models.INDRA database: The INDRA database website provides a search interface to find INDRA Statements assembled from the biomedical literature, browse their supporting evidence, and curate any errors. An example search relevant to COVID-19 is Object: TMPRSS2 to find entities that regulate the TMPRSS2 protease, which is crucial for SARS-CoV-2 entry into human cells.INDRA network search: The INDRA network search allows finding causal paths, shared regulators, and common targets between two entities. An example search relevant to COVID-19 is Subject: ACE2, Object: MTOR (seehere).Dialogue.bio: The dialogue.bio website allows launching dedicated human-machine dialogue sessions where you can upload your data (e.g., DE gene lists or gene expression profiles), discuss relevant mechanisms, and build model hypotheses using simple English dialogue. For instance, you could try the following series of questions: "what is ACE2?", "what does it regulate?", "which of those are transcription factors?".- CLARE is a machine assistant that can be installed in any Slack workspace as
an application. It supports direct messages or messages in channels to
conduct dialogues about biological mechanisms. See demo video
here. It is currently deployed in multiple workspaces and has answered hundreds of questions from COVID-19 researchers since the pandemic began. Pleasecontact usif you would like to install CLARE in your Slack workspace.
This work is funded under the DARPA Communicating with Computers (W911NF-15-1-0544), DARPA Automating Scientific Knowledge Extraction (HR00111990009) and DARPA Automated Scientific Discovery Framework (W911NF-18-1-0124) programs.