- Download zip file
or
git clone https://github.com/phi-grib/RDTExtractorWeb
Install and activate the enviroment
cd RDTExtractorWeb
conda env create -f environment.yml
In Linux, activate the environment using:
source activate RDTExtractorWeb
In Windows, use:
activate RDTExtractorWeb
You will need to put the data files, that are distributed separately, in the (root folder)/API/static/data/ folder.
At the root folder execute:
python manage.py runserver
and then navigate to http:/127.0.0.1:8000.
This tool is designed to extract data from the in vivo repeat-dose toxicity (RDT) studies' database generated within the context of the eTOX project. These data are expanded using an histopathological observation and an anatomical entity ontologies. The histopathological ontology is obtained from Novartis and can be used under the Apache License 2.0. The anatomical entities ontology is extracted from the following paper:
In order to be able to aggregate the data by parent compound, some pre-processing has to be done to data as they exist in the database. Each substance is standardised according to the following protocol:
- From this repository use the process_smiles.std method to standardize, discard mixtures, discard compound with metal ions, and remove all salts. Also use the neutralise.run method to neutralise all charges when possible.
- Using molVS, get the canonical tautomer.
This project is an extension of the work published in the following paper:
Exract studies' findings based on the given filtering and the organs' and morphological changes' ontologies-based expansions of these findings.
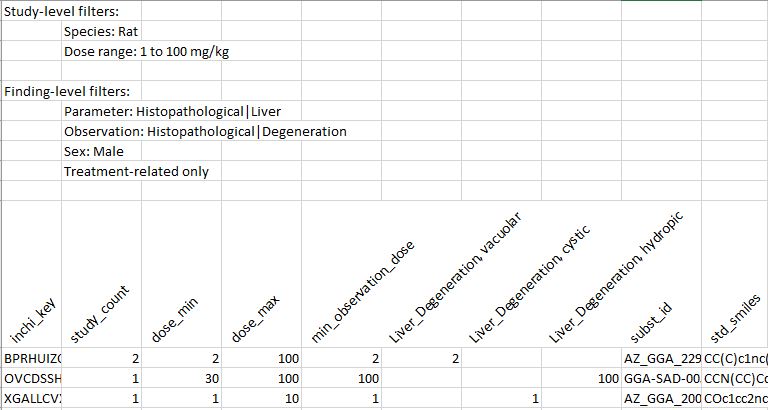
On clicking the 'Extract' button, two output files are generated, one with quantitative and the other with qualitative data. Both have a caption summarising the filtering criteria applied. After this caption, they both have a table with the data aggergated by parent compound. The table contains several fixed columns, namely 6 at the begining:
- inchi_key: Parent compound's InChIKey.
- study_count: Number of relevant studies (according to the current filtering scheme) in which the compound appears.
- dose_min: Minimum dose at which the compound has been tested among the relevant studies.
- dose_max: Maximum dose at which the compound has been tested among the relevant studies.
- min_observation_dose: Minimum dose for which a relevant finding (according to the current filtering scheme) has been reported for the compound.
- is_active: Boolean indicating whether the substance has been found to have any toxicity according to the current finding-related filtering criteria. And two at the end:
- subst_id: All substance IDs corresponding to the parent compound.
- std_smiles: Smiles string corresponding to the standardised parent compound.
Between these two groups, there is a column for each relevant finding. In these columns a value is provided if the finding is reported for the given substance, and it is empty otherwise. The value will be the number of studies that report the finding in the qualitative file, and the minimum dose at which the finding is reported in the quantitative file.