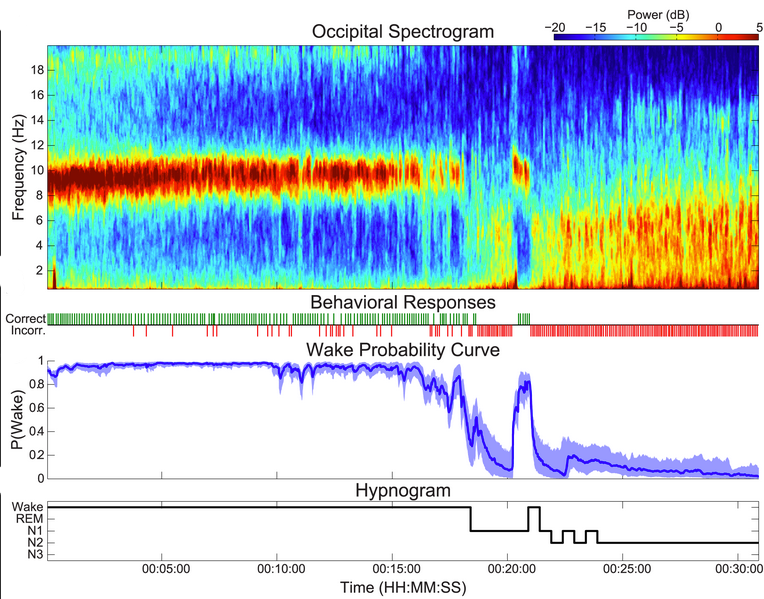
In this repository, we develop new way of tracking the continuum of sleep onset, which incorporates EEG, physiological, and behavioral information to estimate the instantaneous probability that a subject is awake at each point in time.
Prerau, M. J., Hartnack, K. E., Obregon-Henao, G., Sampson, A., Merlino, M., Gannon, K., Bianchi, M., Ellenbogen, J., & Purdon, P. L. (2014). Tracking the sleep onset process: an empirical model of behavioral and physiological dynamics. PLoS computational biology, 10(10), e1003866. https://doi.org/10.1371/journal.pcbi.1003866
How can we tell when someone has fallen asleep? Understanding the way we fall asleep is an important problem in sleep medicine, since sleep disorders can disrupt the process of falling asleep. In the case of insomnia, subjects may fall asleep too slowly, whereas during sleep deprivation or narcolepsy, subjects fall asleep too quickly. Current methods for tracking the wake/sleep transition are time-consuming, subjective, and simplify the sleep onset process in a way that severely limits the accuracy, power, and scope of any resulting clinical metrics. We have developed a new physiologically principled method that dynamically combines information from brainwaves, muscle activity, and a novel minimally-disruptive behavioral task, to automatically create a continuous dynamic characterization of a person’s state of wakefulness.
Specifically, we propose a fully Bayesian dynamic state-space model framework for the characterization of simultaneously observed behavioral and physiological dynamics during the SOP, which is implemented using a partical filter approach. In doing so, we create a robust quantitative representation of SOP dynamics that can be used to more accurately and more precisely track the gradual transition from wakefulness to sleep.
A video tutorial providing an overview of the general concepts underlying the code can be found here.
%ESTIMATE_WAKE_PROB Estimates wake probability from behavioral and physiological data
%
% This code implements the Baysian framework, empirical wake probability model, and particle filter
% described in Prerau et. al 2014, PLOS Computational Biology. Using behavioral (binary responses)
% and physiological (EMG and EEG) data, we can estimate the instantanous probability that a subject
% is awake, which is equivalent in this model to the instantanous probability of response.
%
% Usage:
% estimate_wake_prob() [GENERATES EXAMPLE DATA AND RUNS DEMO]
% parameter_estimates=estimate_wake_prob(Fs, data, num_particles, ploton, prog_bar)
%
% Input:
% Fs: sampling frequency (in Hz)
% data: 5xT matrix of simutaneously observed data, with rows:
% 1. Behavioral response (1=correct, 0=incorrect, NaN=missing at that time point)
% 2. Squeeze amplitude (in mV)
% 3. Alpha power (in dB)
% 4. Delta power (in dB)
% 5. Theta power (in dB)
% All rows are sampled at Fs, with missing data represented by NaN.
% A missing observation type is represented by a complete row of NaN values.
% num_particles: Number of particles to use (Default: 5000)
% ploton: 1 = Plot output graph, 0 = No output plot (Default: 1);
% progbar: 1 = Display progress bar, 0 = No progress bar (Default: 1);
%
% Output:
% estimates: A structure with the 2.5, 50, and 97.5 percentiles of Pr(Wake), observation, and state estimates%Runs demo and saves example data to workspace
parameter_estimates=estimate_wake_prob();