Calculating the fetal fraction for noninvasive prenatal testing based on genome-wide nucleosome profiles
This document is meant to guide you using SANEFALCON. For information on the algorithm itself please refer to the paper this method was published in.
Before you begin...
- Although source code is available, please respect our work and contact us in case of (planned) commercial use.
- Please report bugs through the issue tracker. The implementation was a one man project. Due to time constraints some parts were not well tested before uploading. Mistakes are likely to show up
- I created this hoping it would be useful. If you can't use it, it's not useful. Please let me know of any serious trouble you ran into if you feel there is a mistake on my end.
- I tried my best at making a readme/manual on how to use SANEFALCON. I can imagine it is rather overwhelming. If you do not understand what to do somewhere, let me know so I can update this README or the wiki.
- This implementation is not well done, I know. It's not user friendly, it's difficult to use and it's not as fast as it could be. It's a prototype of the method. If I had enough time I'd love to do a proper (re-)implementation but my current contract does not last forever.
- I created a chat linked to this project on gitter.im. Please use this to discuss this project. Anyone can join using a free GitHub account. Click the link or the badge above to join.
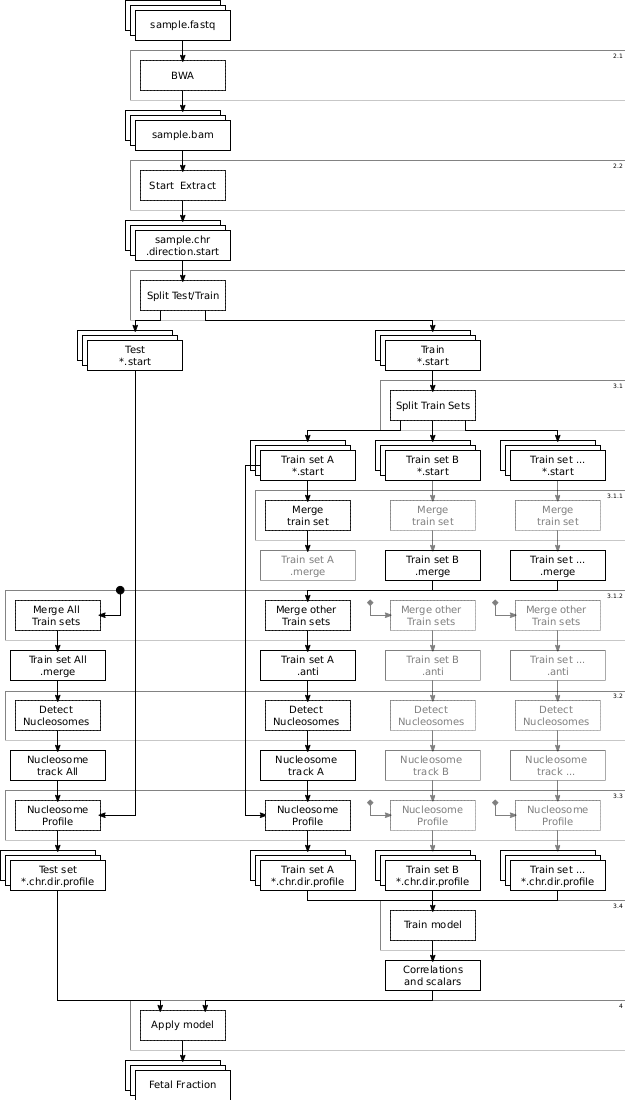
- There is an overview diagram at the bottom of this page. Don't forget to use it to understand where you are and where you should be heading.
- Commands you should enter into a terminal are shown as a code block with a > in front, like so:
./example.sh - In Bash, variables are accessed like this:
$VARIABLE. You are expected to replace such occurrences in this README where needed, or assign values to them when you build your pipeline. - There are many
script.shfiles in this project. Most of them are partial implementations of the pipeline. Study them carefully if you whish to save some time.
Tools used from other projects are shown below. Note that several of the selected software packages could be replaced by alternatives of your choice but were not tested.
- Linux
- BWA
- PicardTools
- SamTools
- Python
--numpy
--scipy
--sklearn
--matplotlib
We used BWA to map our data. To ensure we only have perfect matches we ignore reads that map with either a mismatch or with multiple mappable positions. Before moving on, make sure all data is sorted and preferrably have duplicates removed.
For the technique we applied we are only interested in read start positions. Any information such as the read length etc is neglected as it does not provide us any information. To remove read tower and other side effects of sequencing and mapping we suggest running the data through the RETRO filter as we created for WISECONDOR. In SANEFALCON we supply a slightly altered version to allow extraction of the the read positions. There is no problem replacing this with any method you desire, just be sure to shift the start position for reads mapped to the reverse strand: The BAM format writes the first base pair position of a read. This is not the first in the sequence of the fragment, instead it is the lowest value position the read covers: it is always the leftmost end seen from the reference genome. This is not where the read fragment started, the real position is (the position in the BAM file) + (the length of the read) - 1. The -1 is caused by the fact it is the position covered. We used 51 bp reads, so any read mapped to the reverse strand is actually starting 50 bp downstream, hence the +50 in the script. If you choose to use a different read length, alter this number accordingly. If you use some clipping, introducing variable read lengths, you will have to determine the length per read and use this number instead.
To run the sample extraction, run:
> ./prepSamples.sh $INDIR $OUTDIR
This will attempt to create $OUTDIR, then find any BAM type of file in any subdirectory of $INDIR and process it to extract forward and reverse reads per chromosome, all split over seperate files, i.e.:
$OUTDIR/$SAMPLENAME.1.start.fwd
$OUTDIR/$SAMPLENAME.2.start.fwd
$OUTDIR/$SAMPLENAME.3.start.fwd
...
$OUTDIR/$SAMPLENAME.1.start.rev
$OUTDIR/$SAMPLENAME.2.start.rev
$OUTDIR/$SAMPLENAME.3.start.rev
...
To make sure the nucleosome profiles do not get overtrained it is not possible to apply a nucleosome track to obtain a nucleosome profile if the sample for the latter is already seen in the first. It would influence the called nucleosome positions heavily, causing much stronger pronounced differences between linker and nucleosome covered regions in the profiles. If you have unlimited amounts of data you may skip this step and simply combine all training data for a single nucleosome track, but that means you can never apply the samples used for training in a later step. To reduce the amount of training data required overall it is best to split the data into several batches. In our research, we used about 300 for training in total, split over 12 different batches of about 25 samples each.
- The exact amount of samples per batch is not that important. Roughly the same amount of samples or reads per batch is enough. We mostly chose this to reduce compute time, you may prefer to just split per run itself, accepting varying sample and read counts per subset.
- Do not spread samples from one run over multiple batches, this could influence the nucleosome track by introducing per-run biases providing information for the samples which is not there when testing later on.
- Mixing different runs per batch is a way to approximate the targeted set size. If possible, mix runs from different points in time to reduce any possible batch and time effects from lab work.
- While samples that have an unknown fetal fraction (with female fetus for example) may not provide useful information to fit our model on in a later step, they can be used to determine the most likely nucleosome positions. They should just be ignored later on.
For ease of use and tracking, make a folder structure as shown next:
./test-- contains non training samples./train-- contains only subfolders./train/a-- contains only subfolders, 26 samples total./train/a/run1-- contains 10 samples./train/a/run3-- contains 04 samples./train/a/run6-- contains 12 samples./train/b-- contains only subfolders, 23 samples total./train/b/run2-- contains 16 samples./train/b/run4-- contains 07 samples./train/c/...-- contains only subfolders,...-- ...
The following steps have to be done for each of these subsets, as well as all subsets combined.
Merge start positions per chromosome for every subset of samples you want to combine for a nucleosome track. A standard built-in command for linux is sort:
> sort -n -m files
A little script format to do this is supplied in merge.sh, to run this for subset a use:
> ./merge.sh ./train/a
A loop can save you some manual inputs, replace the letters to match your subdirectories:
> for SUBSET in a b c d e f g h i j k l; do merge.sh ./train/$SUBSET; done
Combine all merged subsets into a merge that covers all subsets as well to create a nucleosome track for testing purposes by running mergeSubs.sh:
> ./mergeSubs.sh ./train
This step needs to be done carefully as it may be confusing: we create an "anti" merge per subset: merge all samples from all subsets except for the subset we want to create a nucleosome track for. For example, set A will get combined read start positions for set B, C, D and so on. Everything EXCEPT set A. Then we do the same thing for set B and so on, this way we will always have a nucleosome track for a subset that was not influenced by the samples in the subset itself.
As we already merged per set previously, we can just merge the subset merges. To do this, run mergeAntiSub.sh for every subset. It contains commands that will collect all merged data except for the targetted subset and merge the other sets per chromosome:
> ./mergeAntiSub.sh ./train $SUBSET
Replace $SUBSET with the subset you want to apply this step for and repeat for every subset you made. Again, a loop can save you some manual inputs, replace the letters to match your subdirectories:
> for SUBSET in a b c d e f g h i j k l; do mergeAntiSub.sh ./train $SUBSET; done
For every set of combined reads a nucleosome track has to be made per chromosome. The script to do this is nuclDetector.py. Most of this script is currently hardcoded and it takes only 2 arguments, input and output. to run for a single subset and chromosome, try something like this:
> python nuclDetector.py $INPUT $OUTPUT
The $INPUT is the merged read start positions for one chromosome (either from a subset or the total set of training samples).
The $OUTPUT is a file with a list of nucleosome positions and their scoring metrics (pos,score,leftVal,innerVal,rightVal), allowing some filtering steps later on.
Now to get a nucleosome track FOR a subset we need to detect nucleosomes on the "anti" merged data for further processing. To run for a subset use nuclDetectorAnti.sh:
> nuclDetectorAnti.sh ./train/$SUBSET
Or, again, use a loop:
> for SUBSET in a b c d e f g h i j k l; do nuclDetectorAnti.sh ./train/$SUBSET; done
Also, create a nucleosome track for the whole set of training data:
> ./nuclDetector.sh ./train
At this point it is probably a good idea to check whether all of your nucleosome tracks are slightly different from eachother: It's pretty much impossible for all of them to find the exact same nucleosome positions, nor do they differ hugely. Most positions should be close to eachother over different files, and pretty much every position will be the same over a few different tracks. Just open a few and look into the first 10 to 20 nucleosome calls and see if this behaviour is there. If not, you may have applied the nucleosome detection several times on the exact same data.
Next every sample in the training set is analyzed to obtain an aligned nucleosome profile. The script to do this is getProfile.py and needs to be run 4 times per chromosome per sample. The reason for this is it needs to take all reads into account in both directions, both upstream and downstream from the nucleosome positions, and this happened to be easier to write.
Arguments are:
$NUCL.$CHROM: Nucleosome track file for this chromosome. Be aware: for a training sample, use the nucleosome track that is based on the "anti" merge for its subset, hence the whole preparation up to this point. DO NOT APPLY A NUCLEOSOME TRACK WHERE THE SAMPLE CAN HAVE INFLUENCED THE NUCLEOSOME CALLS$SAMPLE.$CHROM.start.fwd: The file containing read start positions. These are split by forward and reverse stranded reads, hence the fwd and rev parts in the names.0/1: The direction the script looks for reads relative to the nucleosome center. 0 is upstream, 1 is downstream. Using a 1 on a fwd track means you are looking at reads starting AFTER the nucleosome center. The same goes for using a 0 on a rev track.$SAMPLEOUT.$CHROM.fwd/ifwd/rev/irev: The output file for the determined profile. Save every output in a separate file, do not overwrite or combine yet.
As an example, arguments are shown here as variables:
> python getProfile.py $NUCL.$CHROM $SAMPLE.$CHROM.start.fwd 0 $SAMPLEOUT.$CHROM.fwd
> python getProfile.py $NUCL.$CHROM $SAMPLE.$CHROM.start.fwd 1 $SAMPLEOUT.$CHROM.ifwd
> python getProfile.py $NUCL.$CHROM $SAMPLE.$CHROM.start.rev 1 $SAMPLEOUT.$CHROM.rev
> python getProfile.py $NUCL.$CHROM $SAMPLE.$CHROM.start.rev 0 $SAMPLEOUT.$CHROM.irev
While this provides a profile per chromosome, per direction, per sample, we only want a single profile per sample. To do this, we will combine all downstream profiles over all chromosomes per sample in one array and all upstream profiles per sample in another, then merge the two together to create a full length read start profile that covers the whole region surrounding nucleosome positions.
The following scripts will combine the read start profiles per direction:
> ./combProf.sh searchDir >> upstreamProfs.csv
> ./combProfI.sh searchDir >> downstreamProfs.csv
These scripts will use a built-in find function to detect profile files and merge them per upstream or downstream direction for each sample. At this point, I do not have a seperate script to combine the upstream and downstream profiles. Instead just open them in a spreadsheet tool, sort them both by file name so their orders are aligned and reverse the upstream profile (left most value must become rightmost etc), then stitch the two together by putting the downstream profiles after the upstream profiles. Note there is a value in the middle that has to be removed as both sides also take the nucleosome center base pair into account, thus providing this number twice. You should find the last number from the upstream matches the first number of the downstream profiles when putting them together. Just delete one of the two columns to get rid of this problem, then save the combined profiles in a new file (plain text/csv) for further analysis. The final result should contain the samplename in the first column, then followed by only read start counts per position in every column after that.
Use the python script as follows to see how far we've come and train a simple convertor for the future:
> python predictor.py trainNucl trainRef outBaseName
This script has 3 main and 2 optional arguments:
trainNuclTraining nucleosome profilestrainRefTraining reference dataoutBaseNameOutput basename
s1. Test nucleosome profiles
s2. Test reference data
If supplied, it will directly analyze your test samples if they were provided in the same formats as the training samples (arguments s1 and s2). The script will match sample names in nucleosome profile files with names in the reference files, taking the first column for each as containing the names. The reference data should contain the estimated fetal fraction based on an existing method in the second column, for example the chromosome Y frequency for male pregnancies. The output will provide a plot showing the correlation profile and a scatterplot with the estimated fraction using SANEFALCON vs the reference values where available.
All steps for test samples have been described in the training steps, but now the subset splitting and nucleosome detection steps can be skipped:
- Obtain read start positions
- Get nucleosome profiles, now use the profile based on all training samples (subsets) combined
- Feed the new data as argument 4 in the final training phase, 5th argument should be fractions obtained in the same way as for the training reference to check the estimation quality of SANEFALCON. If not available just point to the same file as the second argument, as a result no samples will match so SANEFALCON will assume these samples are female and thus unknown.
This method is easier and was created later on. This allows implementation in pipelines for day to day use. The original method is kept as it provides insight on the data and algorithm.
If you trained the model previously, it created a outBaseName.model file. This file describes how the nucleosome profile correlates with the fetal fraction and what linear formula to use to shift its output to the same scale as the original input reference fetal fraction data.
There is a shell script that prepares the nucleosome profiles for a small python script:
> predict.sh trainedModel testProfile
where:
trainedModelis theoutBaseName.modelfiletestProfileis the basename of files that contain the forward and reverse nucleosome profile per chromosome for a single test sample.
It will add nucleosome profiles per strand into a single file and apply the model to turn this into an estimate of the fetal fraction. Output is saved into testProfile.ff.
Overview of the whole SANEFALCON script. Small numbers in the top right of boxes are the subsections in this document where the step is described.