-
Notifications
You must be signed in to change notification settings - Fork 0
Iteration 5 Architecture
This wiki page describes the architecture for Iteration 5 of the SCOOP project.
The architecture context is basically the same as the previous iteration; however, we are enhancing and testing the hQuery components.

This iteration is being deployed to a laboratory environment (non production) at UVic.
There are two practices being simulated in this iteration (i.e. two endpoints) via discrete virtual machines.
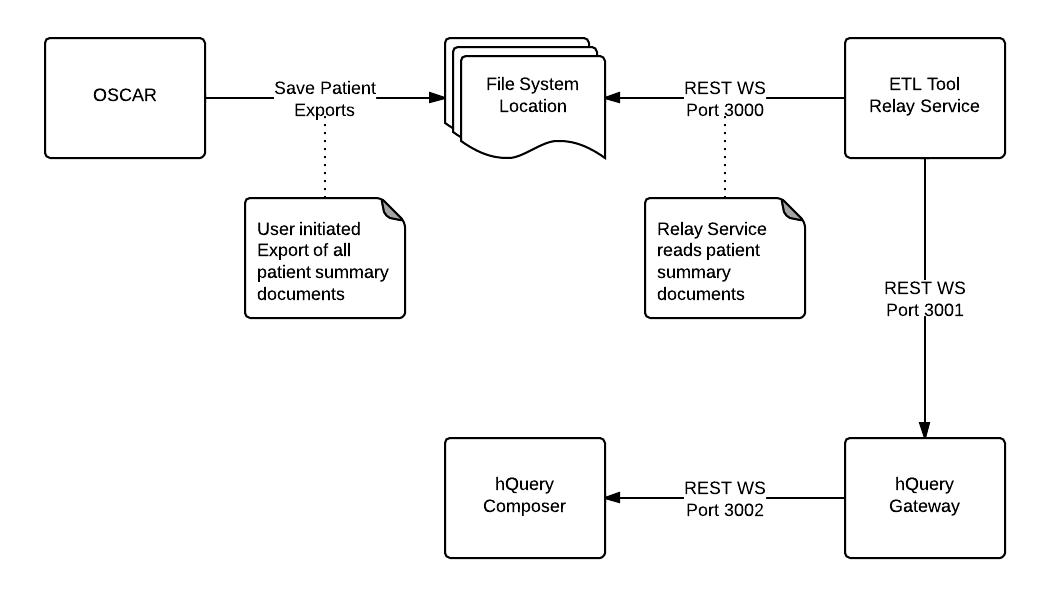
There are a number of architectural changes planned for this iteration of SCOOP.
1. We are continuing to enhance and refactor OSCAR to export the patient summary document, either for a single individual or by batch for all individuals with a record in the EMR
2. The middleware (relay-service.rb) A simple ruby webservice will transfer individual summary documents between the EMR and the hQuery Gateway. It will not perform any aggregation or transformation.
3. hQuery Hub will be investigated as to how we can extend the reporting functionality of the resultant queries. This will allow for future iterations to have a more meaningful result for the researcher by providing a better view of the information.
Privacy Individual information never leaves the practice. Early feedback from discussions with the faculty physicians UBC highlighted their stewardship of patient information as a serious matter. There’s a genuine concern that anonymous data collected may be used for unintended research purposes in the future. Several are concerned by the risks of data aggregation, even when the data is de-identified.
Open Source As SCOOP development is sponsored with public funds it will be released as open source for anyone to use. In addition, SCOOP will only rely on free, open source components so that its use is accessible to any individual/group.
OSCAR In addition to being open source, OSCAR is the EMR used by many UBC clinical faculty and physicians. A few of these faculty members will be the initial SCOOP users. For this reason, OSCAR will be the first supported EMR.
Software design of the export feature in OSCAR
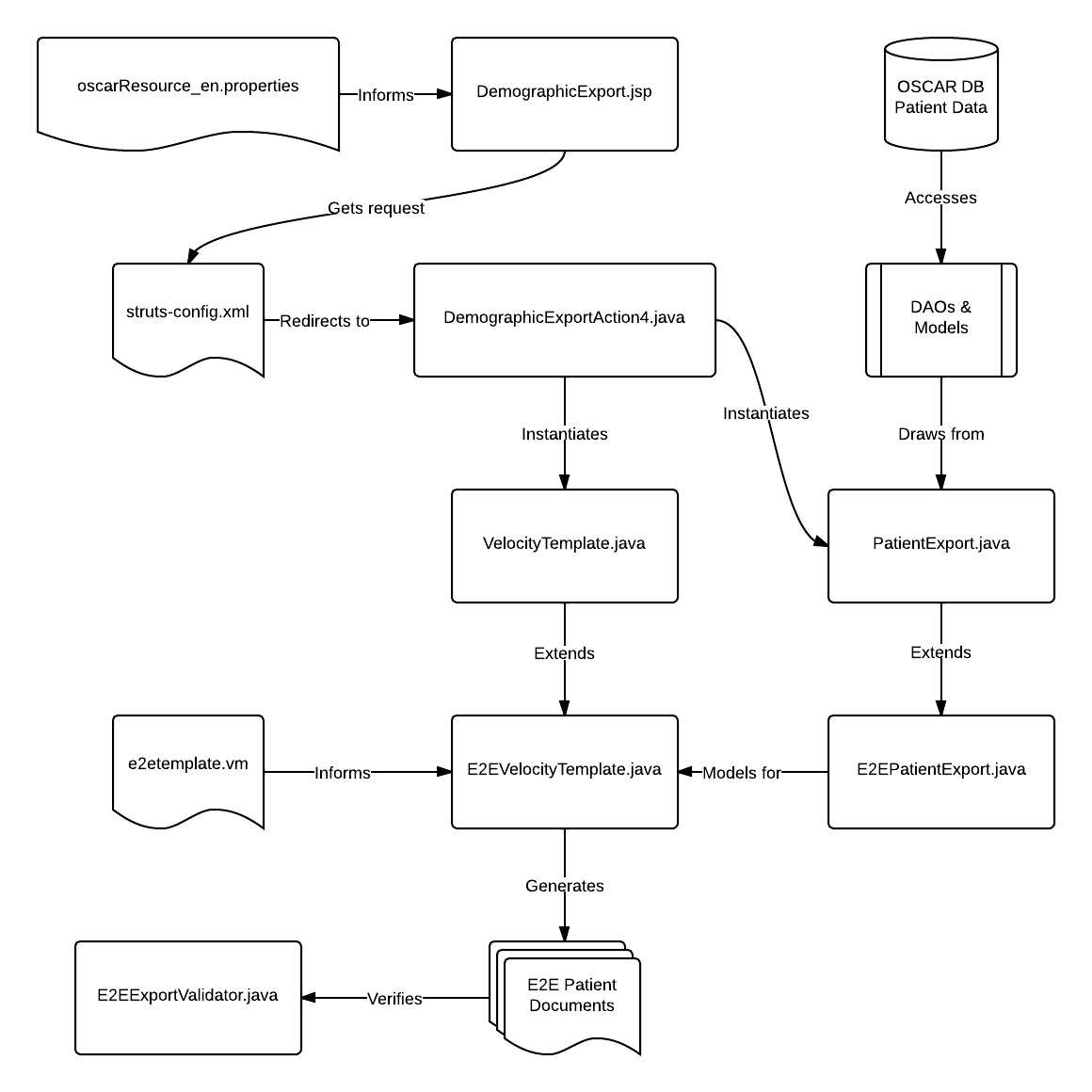
We will attempt to stay true to the MVC design. Velocity based templating requires both a template and a data model. We have the PatientExport object draw on all the pre-existing DAOs and models and have it group up the relevant fields for easy velocity access. Most of the heavy code work will be found in e2etemplate.vm, and DemographicExportAction4.java acts as the Event Handler and file management for the export function.
Software design for new hQuery enhancements
Our question draws on demographics, medications and laboratory results; all of which are supported by hQuery's API. There shouldn't be any changes to hQuery's API this iteration. However, we will be migrating all of hQuery and its dependencies to Ruby 1.9.3 because of the security vulnerabilities found in 1.9.2 as well as clean up the dependency trees that hQuery has.
Query Design
Since hQuery follows the map-reduce paradigm for gathering and returning results, we will also be using a mapping and reducing design. Since Iteration 5 is a non-functional update, it is expected that all previous queries will still be able to function correctly.
Since Iteration 5 is a non-functional update, we expect all previous queries to function correctly. As well, it was also our goal to include the original scope of IT4's query which includes dosage and lab values. The new map function with those features are added below.
function map(patient) {
var targetMedicationCodes = {
"whoATC": ["C01AA05"],
"HC-DIN": ["02281236", "02281228", "02281201", "02245428", "02245427",
"02245426", "02048264", "02048272", "0021415", "00698296", "00647470"]
};
var targetLabCodes = {
"LOINC": ["45066-8", "2160-0", "33914-3", "50044-7", "48642-3", "48643-1"]
};
var ageLimit = 65;
var creatinineLimit = 150; // Measured in umol/L
var digoxinLimit = .125; // Measured in MG
var drugList = patient.medications();
var resultList = patient.results();
var now = new Date(2013, 5, 12);
var start = new Date(2000, 6, 1);
var end = addDate(now, 0, 1, 0);
// Shifts date by year, month, and date specified
function addDate(date, y, m, d) {
var n = new Date(date);
n.setFullYear(date.getFullYear() + (y || 0));
n.setMonth(date.getMonth() + (m || 0));
n.setDate(date.getDate() + (d || 0));
return n;
}
// Checks if patient is older than ageLimit
function population(patient) {
return (patient.age(now) >= ageLimit);
}
// Checks for Creatinine labs performed within the last year
function hasLabCode() {
return resultList.match(targetLabCodes, addDate(now, -1, 0, 0), end).length;
}
// Checks for existence of Digoxin
function hasMedication() {
return drugList.match(targetMedicationCodes, start, end).length;
}
// Checks if Creatinine meets parameters
function hasMatchingLabValue() {
for (var i = 0; i < resultList.length; i++) {
if (resultList[i].values()[0].units() == "umol/L") {
if (resultList[i].values()[0].scalar() > creatinineLimit) {
//emit("Abnormal Creatinine: " + patient.given() + " " + patient.last(), 1);
return true;
}
}
}
return false;
}
// Checks if existing Digoxin is current
function hasCurrentMedication() {
for (var i = 0; i < drugList.length; i++) {
var tmpArray = new hQuery.CodedEntryList();
tmpArray[0] = drugList[i];
if (tmpArray.match(targetMedicationCodes)) {
var drugStart = drugList[i].indicateMedicationStart().getTime();
var drugEnd = drugList[i].indicateMedicationStop().getTime();
// Check if drug is within the right time
if (drugEnd >= now && drugStart <= now) {
return true;
}
}
}
return false;
}
// Checks if Digoxin meets dosage parameters
function hasMatchingMedicationDose() {
for (var i = 0; i < drugList.length; i++) {
var codes = drugList[i].medicationInformation().codedProduct();
// If Digoxin, check for dose parameter
for (var j = 0; j < codes.length; j++) {
if (codes[j].includedIn(targetMedicationCodes)) {
if (drugList[i].values()[0].units() == "MG") {
if (drugList[i].values()[0].scalar() > digoxinLimit) {
return true;
}
}
}
}
}
}
emit('total_pop', 1);
if (population(patient)) {
//emit("senior_pop: " + patient.given() + " " + patient.last(), 1);
emit("senior_pop", 1);
if (hasLabCode() && hasMedication()) {
//emit("Digoxin: " + patient.given() + " " + patient.last(), 1);
//emit("Creatinine: " + patient.given() + " " + patient.last(), 1);
if (hasMatchingLabValue() && hasCurrentMedication() && hasMatchingMedicationDose()) {
emit("senior_pop_digoxin_creatinine", 1);
}
}
}
}
Since our reduction function works for a wide range of inputs, it will remain the same as previous iterations.
function reduce(key, values) {
var result = 0;
while (values.hasNext()) {
result += values.next();
}
return result;
}
Introduction to E2E E2E, or EMR to EMR, is a CDA specification being developed by BC's PITO organization. Its main purpose is to create a standard in which EMR data can be transmitted to other Canadian EMRs in a standardized way.
Overview of E2E sections (including image)
Scope of content for Iteration 5 In order to sufficiently return value back to the OSCAR community, we need to implement a minimally functional full E2E exporter. As such, our scope includes all sections and content that is defined as required by the E2E specification.
OSCAR An Open Source EMR system used by many private physicians. This is way that individual information is captured - and the user interface for the physician practioner end user.
relay-service.rb This is an open source ruby script developed by the SCOOP project. Its main purpose is to allow E2E documents stored within a specified directory to be passed into the query gateway by restful web service calls.
hQuery Gateway The query gateway is a web based application that provides the back end for executing queries. The query gateway which exposes a query API, accepts queries, runs those queries against the patient data, and returns the results of the query back to the query composer.
hQuery Composer The query composer is a web based application that provides the front end for creating, managing, and executing queries. Those queries are executed against the query gateway which exposes a query API, accepts queries, runs those queries against the patient data, and returns the results of the query back to the query composer.
Scoop Endpoint refers to all the non-EMR SCOOP software components that will reside at a practice in the future. This includes the hQuery Gateway and relay-service.rb.
SCOOP Hub This is the application that the researcher uses to ask a "question" of the Research Network. This includes features for query management, policy enforcement, privacy management, and security. Currently, hQuery composer is the only software system in the hub - but more design and enhancements are needed.
- Libraries: template
SCOOP is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License.
- SCOOP Overall Design
- SCOOP Actors
- User Stories and Use Case Maps
- System Architecture
- Development Process
- Prototypical Questions
- Current Meds vs Med List
- Data Enrichment Design
- Data Visualization
- Deployment Architecture
- EMR-2-EMR (E2E)
- OSCAR Setup
- Gateway & Hub Setup
- OSCAR Development Notes
- OSCAR DB Table Notes
- Coding Standards
- Mongodb Notes
- Server Configuration
- PDC Gateway Server
- Iteration Overview
- Feature List
- Architecture
- Requirements
- Visualization Requirements
- Test Specification
